Ubp2 modulates DJ-1-mediated redox-dependent mitochondrial dynamics in Saccharomyces cerevisiae
- PMID: 40608842
- PMCID: PMC12251144
- DOI: 10.1371/journal.pgen.1011353
Ubp2 modulates DJ-1-mediated redox-dependent mitochondrial dynamics in Saccharomyces cerevisiae
Abstract
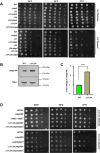
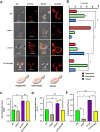
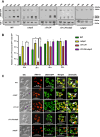
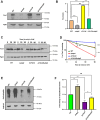
Mitochondrial integrity is a crucial determinant of overall cellular health. Mitochondrial dysfunction and impediments in regulating organellar homeostasis contribute majorly to the pathophysiological manifestation of several neurological disorders. Mutations in human DJ-1 (PARK7) have been implicated in the deregulation of mitochondrial homeostasis, a critical cellular etiology observed in Parkinson's disease progression. DJ-1 is a multifunctional protein belonging to the DJ-1/ThiJ/PfpI superfamily, conserved across the phylogeny. Although the pathophysiological significance of DJ-1 has been well-established, the underlying molecular mechanism(s) by which DJ-1 paralogs modulate mitochondrial maintenance and other cellular processes remains elusive. Using Saccharomyces cerevisiae as the model organism, we unravel the intricate mechanism by which yeast DJ-1 paralogs (collectively called Hsp31 paralogs) modulate mitochondrial homeostasis. Our study establishes a genetic synthetic interaction between Ubp2, a cysteine-dependent deubiquitinase, and DJ-1 paralogs. In the absence of DJ-1 paralogs, mitochondria adapt to a highly tubular network due to enhanced expression of Fzo1. Intriguingly, the loss of Ubp2 restores the mitochondrial integrity in the DJ-1 deletion background by modulating the ubiquitination status of Fzo1. Besides, the loss of Ubp2 in the absence of DJ-1 restores mitochondrial respiration and functionality by regulating the mitophagic flux. Further, Ubp2 deletion makes cells resistant to oxidative stress without DJ-1 paralogs. For the first time, our study deciphers functional crosstalk between Ubp2 and DJ-1 in regulating mitochondrial homeostasis and cellular health.
Copyright: © 2025 Biswas, D’Silva. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

