Unraveling the genetics of feline hypertrophic cardiomyopathy: a multiomics study of 138 cats
- PMID: 40609041
- PMCID: PMC12405878
- DOI: 10.1093/g3journal/jkaf153
Unraveling the genetics of feline hypertrophic cardiomyopathy: a multiomics study of 138 cats
Abstract
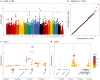
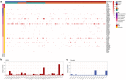
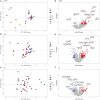
Hypertrophic cardiomyopathy (HCM) is the most common inherited cardiac disease in cats, often leading to congestive heart failure, arterial thromboembolism, and sudden cardiac death. The genetics of feline HCM are poorly understood and limited genetic discoveries remain breed- or family-specific. We aimed to identify novel causative or disease-modifying variants in a large cohort of cats reflective of the general cat population. In a second cohort, we sought to characterize transcriptomics differences between HCM-affected cats and healthy controls. DNA was isolated from 138 domestic cats (109 HCM and 29 controls). No single or combination of variants of high, moderate, or modifying impact were identified by genome-wide analysis to cause or modify disease severity of HCM. Several rare high and moderate impact variants in genes associated with human HCM were detected in diseased cats. In a second cohort, left ventricular (LV), interventricular septal (IVS), and left atrial (LA) tissues of 27 HCM-affected and 15 control cats were submitted for stranded mature RNA-sequencing at 50 million reads/sample. A total of 74, 115, and 45 differentially expressed genes (DEGs) were upregulated and 8, 53, and 48 DEGs were downregulated in LV posterior wall, IVS, and LA tissue, respectively, in HCM-affected cats compared to controls. Similar to humans, the genetic etiology of feline HCM remains unknown in a high proportion of cases. Transcriptomics revealed molecular signatures that may help identify novel HCM biomarkers or drug targets in future investigations.
Keywords: RNA-sequencing; cardiovascular; precision medicine; translational; whole-genome sequencing.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures


References
-
- Agrawal V et al. 2019. Natriuretic peptide receptor C contributes to disproportionate right ventricular hypertrophy in a rodent model of obesity-induced heart failure with preserved ejection fraction with pulmonary hypertension. Pulm Circ. 9:2045894019878599. doi: 10.1177/2045894019895452. - DOI - PMC - PubMed
-
- Animal Welfare Act as Amended. 2013 USC 7:S2131-2159.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
