AI-enabled molecular phenotyping and prognostic predictions in lung cancer through multimodal clinical information integration
- PMID: 40609537
- PMCID: PMC12281446
- DOI: 10.1016/j.xcrm.2025.102216
AI-enabled molecular phenotyping and prognostic predictions in lung cancer through multimodal clinical information integration
Abstract
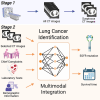
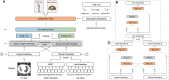
Lung cancer remains the leading cause of cancer-related mortality worldwide. The need for cost-effective, non-invasive methods to detect specific gene mutations for targeted therapy and predict patient survival outcomes underscores the importance of advancing diagnostic and prognostic capabilities. Contemporary lung cancer diagnostic models often fail to integrate diverse patient data, leading to incomplete clinical assessments. To address these challenges, we propose LUCID, a multimodal data integration framework designed to predict epidermal growth factor receptor (EGFR) mutation status and survival outcomes in patients with lung cancer. Tailored for early-stage clinical assessment, LUCID leverages lung computed tomography (CT) images, chief complaints, laboratory test results, and demographic data to deliver comprehensive, non-invasive predictions. LUCID achieved strong performance in a retrospective cohort of 5,175 patients, with areas under the receiver operating characteristic curve (AUCs) ranging from 0.851 to 0.881 for EGFR mutation prediction and from 0.821 to 0.912 for survival time prediction. The model also demonstrated robustness across external validation cohorts and in scenarios with missing modalities.
Keywords: EGFR mutation; lung cancer; multimodal integration; non-invasive diagnosis; precision medicine; survival prediction.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R.L., Soerjomataram I., Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024;74:229–263. - PubMed
-
- Siegel R.L., Giaquinto A.N., Jemal A. Cancer statistics, 2024. CA Cancer J. Clin. 2024;74:12–49. - PubMed
-
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically Applicable AI System for Accurate Diagnosis, Quantitative Measurements, and Prognosis of COVID-19 Pneumonia Using Computed Tomography. Cell. 2020;181:1423–1433.e11. doi: 10.1016/j.cell.2020.04.045. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

