Epigenetic profiling identifies markers of endocrine resistance and therapeutic options for metastatic castration-resistant prostate cancer
- PMID: 40609538
- PMCID: PMC12281433
- DOI: 10.1016/j.xcrm.2025.102215
Epigenetic profiling identifies markers of endocrine resistance and therapeutic options for metastatic castration-resistant prostate cancer
Abstract
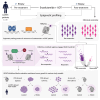
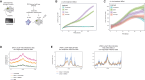
Androgen receptor (AR) signaling inhibitors, including enzalutamide, are treatment options for patients with metastatic castration-resistant prostate cancer (mCRPC), but resistance inevitably develops. Using metastatic samples from a prospective phase 2 clinical trial, we epigenetically profile enhancer/promoter activities with acetylation of lysine residue 27 on histone 3 (H3K27ac) chromatin immunoprecipitation followed by sequencing, before and after AR-targeted therapy. We identify a distinct subset of H3K27ac-differentially marked regions that are associated with treatment responsiveness, which we successfully validate in mCRPC patient-derived xenograft (PDX) models. In silico analyses reveal histone deacetylase (HDAC)3 to critically drive resistance to hormonal interventions, which we validate in vitro. Critically, we identify the pan-HDAC inhibitor vorinostat to be effective in decreasing tumor cell proliferation, both in vitro and in vivo. Moreover, we uncover evidence for HDAC3 working together with glucocorticoid receptor (GR) as a potential mechanism for this therapeutic effect. These findings demonstrate the rationale for therapeutic strategies including HDAC inhibitors to improve patient outcome in advanced stages of mCRPC.
Keywords: H3K27ac; HDAC inhibitors; androgen receptor; biomarkers; drug resistance; enzalutamide; epigenetics; hormone intervention; mCRPC; prostate cancer.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.Z. and A.M.B. received research funding from Astellas Pharma for the work performed in this manuscript. P.S.N. has served as a paid consultant to AstraZeneca, Janssen, Pfizer, and Genentech and received research support from Janssen for work unrelated to the present study.
Figures




Update of
-
Enhancer profiling identifies epigenetic markers of endocrine resistance and reveals therapeutic options for metastatic castration-resistant prostate cancer patients.medRxiv [Preprint]. 2023 Feb 24:2023.02.24.23286403. doi: 10.1101/2023.02.24.23286403. medRxiv. 2023. Update in: Cell Rep Med. 2025 Jul 15;6(7):102215. doi: 10.1016/j.xcrm.2025.102215. PMID: 36865297 Free PMC article. Updated. Preprint.
References
-
- Mottet N., van den Bergh R.C.N., Briers E., Van den Broeck T., Cumberbatch M.G., De Santis M., Fanti S., Fossati N., Gandaglia G., Gillessen S., et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021;79:243–262. doi: 10.1016/j.eururo.2020.09.042. - DOI - PubMed
-
- Kupelian P.A., Mahadevan A., Reddy C.A., Reuther A.M., Klein E.A. Use of different definitions of biochemical failure after external beam radiotherapy changes conclusions about relative treatment efficacy for localized prostate cancer. Urology. 2006;68:593–598. doi: 10.1016/j.urology.2006.03.075. - DOI - PubMed
-
- Roehl K.A., Han M., Ramos C.G., Antenor J.A.V., Catalona W.J. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3,478 consecutive patients: long-term results. J. Urol. 2004;172:910–914. doi: 10.1097/01.ju.0000134888.22332.bb. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

