The mechanistic basis for interprotomer deglycosylation of antibodies by corynebacterial IgG-specific endoglycosidases
- PMID: 40610417
- PMCID: PMC12229479
- DOI: 10.1038/s41467-025-60986-w
The mechanistic basis for interprotomer deglycosylation of antibodies by corynebacterial IgG-specific endoglycosidases
Abstract
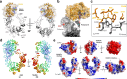
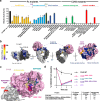
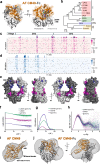
Corynebacterium diphtheriae clade species secrete single-domain endo-β-N-acetylglucosaminidases (ENGases) that specifically bind to human IgG antibodies and hydrolyze their N297-linked glycans. Here, we define the molecular mechanisms of IgG-specific deglycosylation for the entire family of corynebacterial IgG-specific ENGases, including but not limited to CU43 and CM49. By solving the crystal structure of CU43 in a 1:1 complex with the IgG1 Fc region, combined with targeted and saturation mutagenesis analysis and activity measurements using engineered antibodies, we establish an inter-protomeric mechanism of recognition and deglycosylation of IgG antibodies. Using in silico modeling, small-angle X-ray scattering and saturation mutagenesis we determine that CM49 uses a unique binding site on the Fc region, to process N297-linked glycans. Moreover, we demonstrate that CU43 treatment is highly effective in abrogating Fc effector functions in humanized mouse models, while preserving the neutralizing capacity of anti-influenza IgG antibodies, thereby conferring protection against lethal influenza challenge.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.E.S., S.B, J.D., J.V.R. and E.J.S. are inventors on a provisional patent application filed with the United States Patent and Trademark Office by Emory University relevant to the work in this manuscript. All other authors declare they have no competing interests.
Figures





References
-
- Fairbanks, A. J. The ENGases: versatile biocatalysts for the production of homogeneous N-linked glycopeptides and glycoproteins. Chem. Soc. Rev.46, 5128–5146 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

