Clinical and metabolic consequences of a historic pathogenic lamin A/C founder variant
- PMID: 40610603
- PMCID: PMC12229498
- DOI: 10.1038/s41598-025-08495-0
Clinical and metabolic consequences of a historic pathogenic lamin A/C founder variant
Abstract
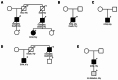
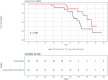
A novel LMNA p.(Glu105Leu) variant was identified in five families with dilated cardiomyopathy (DCM), revealed as a local founder variant originating approximately 650 years ago. Genetic testing and clinical analysis of 795 DCM patients demonstrated that probands with this variant typically present with severe DCM in their sixties, characterized by high prevalence of late gadolinium enhancement, arrhythmias, and conduction disorders. Time-to-event analysis suggested a later onset of clinical symptoms compared to other LMNA variants, with a trend towards longer event-free survival. Microscopic imaging of patient fibroblasts, induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs), and heart tissue confirmed structural nuclear LMNA-associated abnormalities. Patient iPSC-CMs exhibited distinct sarcomeric disorganization, increased glucose uptake and glycogen content, reduced mitochondrial function and biogenesis, and delayed contractile function. These findings support the pathogenicity of the variant and demonstrate its profound impact on structural and metabolic functions in cardiomyocytes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Heymans, S., Lakdawala, N. K., Tschöpe, C. & Klingel, K. Dilated cardiomyopathy: causes, mechanisms, and current and future treatment approaches. Lancet402, 998–1011. 10.1016/S0140-6736(23)01241-2 (2023). - PubMed
-
- Weintraub, R. G., Semsarian, C. & Macdonald, P. Dilated cardiomyopathy. Lancet390, 400–414. 10.1016/S0140-6736(16)31713-5 (2017). - PubMed
-
- Verdonschot, J. A. J. et al. Implications of genetic testing in dilated cardiomyopathy. Circ. Genom Precis Med.13, 476–487. 10.1161/circgen.120.003031 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

