Cancer cell type-specific derepression of transposable elements by inhibition of chromatin modifier enzymes
- PMID: 40610675
- PMCID: PMC12229592
- DOI: 10.1038/s42003-025-08413-0
Cancer cell type-specific derepression of transposable elements by inhibition of chromatin modifier enzymes
Abstract
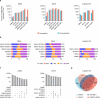
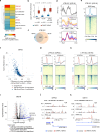
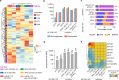
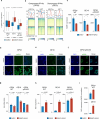
Derepression of transposable elements (TE) by epigenetic therapy leads to the activation of immune response in cancer cells. However, the molecular mechanism of TE regulation by distinct chromatin modifier enzymes (CME) in context of p53 is still elusive. Here, we used FDA-approved epigenetic drugs to systematically inhibit distinct CMEs in p53 wild-type and p53-mutant colorectal, esophageal, and prostate cancer cells. We show that distinct TE subfamilies are derepressed by inhibition of different CMEs in cell type-specific manner. Co-inhibition of DNMT and HDAC (DNMTi-HDACi) had the most consistent effect across cancer types. Loss of p53 results in stronger TE activation and TE-chimeric transcript expression and this effect is largely mediated by the non-genomic actions of p53. Robust immune response elicited by DNMTi-HDACi is due to induced inverted repeat Alu expression concomitant with reduced ADAR1-mediated Alu RNA editing. Collectively, our systematic analyses provide insights for rational use of epigenetic therapies in distinct cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Epigenetic editing and epi-drugs: a combination strategy to simultaneously target KDM4 as a novel anticancer approach.Clin Epigenetics. 2025 Jun 19;17(1):105. doi: 10.1186/s13148-025-01913-0. Clin Epigenetics. 2025. PMID: 40537846 Free PMC article.
-
Multi-Cohort Exploration of Repetitive Element Transcription and DNA Methylation in Human Steatotic Liver Disease.Int J Mol Sci. 2025 Jun 8;26(12):5494. doi: 10.3390/ijms26125494. Int J Mol Sci. 2025. PMID: 40564961 Free PMC article.
-
Concurrent RB1 and P53 pathway disruption predisposes to the development of a primitive neuronal component in high-grade gliomas depending on MYC-driven EBF3 transcription.Acta Neuropathol. 2025 Jan 16;149(1):8. doi: 10.1007/s00401-025-02845-y. Acta Neuropathol. 2025. PMID: 39821672
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
A narrative review of epigenetic marker in H3K27ac and its emerging potential as a therapeutic target in cancer.Epigenomics. 2025 Mar;17(4):263-279. doi: 10.1080/17501911.2025.2460900. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39981972 Review.
References
-
- Burns, K. H. Transposable elements in cancer. Nat. Rev. Cancer17, 415–424 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

