This is a preprint.
Complete cross strain protection against congenital cytomegalovirus infection requires a vaccine encoding key antibody (gB) and T-cell (immediate early 1 protein) viral antigens
- PMID: 40611892
- PMCID: PMC12224539
- DOI: 10.1101/2025.06.18.660432
Complete cross strain protection against congenital cytomegalovirus infection requires a vaccine encoding key antibody (gB) and T-cell (immediate early 1 protein) viral antigens
Update in
-
Complete cross strain protection against congenital cytomegalovirus infection requires a vaccine encoding key antibody (gB) and T-cell (immediate early 1 protein) viral antigens.Front Immunol. 2025 Oct 22;16:1649656. doi: 10.3389/fimmu.2025.1649656. eCollection 2025. Front Immunol. 2025. PMID: 41200170 Free PMC article.
Abstract
Background: Cytomegalovirus is a leading cause of congenital disease and multiple strains enable congenital CMV (cCMV) from both primary and non-primary infection. A cross-strain protective cCMV vaccine is a high priority. The guinea pig is the only small animal model for cCMV and guinea pig cytomegalovirus (GPCMV) encodes functional homolog proteins including cell entry gB glycoprotein and non-structural immediate early 1 protein (IE1), essential for lytic infection. A gB vaccine antibody response fails to provide horizontal protection against highly cell-associated clinical GPCMV strain TAMYC compared to prototype strain 22122. Previously, a recombinant defective adenovirus (Ad) vaccine encoding IE1, a T-cell antigen, provided high-level cCMV protection. In this study, we hypothesized that a combined Ad-based strategy encoding trimeric gB complex and IE1 (AdgB+AdIE1) could improve cross-strain protection against cCMV compared to a gB vaccine (AdgB).
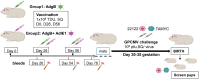
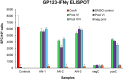
Methods: A preconception vaccine study evaluated the immune response and ability of vaccines to provide cross-strain protection against cCMV. Seronegative female animals were assigned into three vaccine groups: Group 1 (AdgB); Group 2 (AdgB+AdIE1); Group 3 (no vaccine). Animals were vaccinated following a previously defined protocol and antibody ELISAs were used to evaluate gB immune response (AD1, prefusion gB and wild type gB). Additionally, an IFNγ-ELISPOT assay evaluated IE1 T-cell response. During second trimester dams were challenged with GPCMV (22122 and TAMYC) and pregnancy went to term where viral loads in pup target organs and placentas were evaluated.
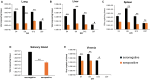
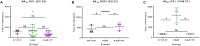
Results: Vaccinated dams elicited a higher neutralizing antibody response to gB than natural convalescent immunity and antibodies recognized homolog AD1 gB domain as well as prefusion gB with response surpassing natural immunity. Group 2 animals additionally elicited a T-cell response to IE1. Evaluation of viral load in pups demonstrated that AdgB+AdIE1 vaccine reduced GPCMV transmission to below detectable limits compared to 91.7% in unvaccinated group. In contrast, AdgB reduced cCMV transmission to 12% in pups.
Conclusion: Complete cross-strain cCMV protection is a significant milestone in this model and achieved by inclusion of an antibody response to trimeric gB and T-cell response to IE1. Importantly, gB and IE1 responses can synergize and increase protection against cCMV unlike prior approaches.
Keywords: CMV vaccine; IE1; congenital CMV; cytomegalovirus; gB; guinea pig; placenta.
Conflict of interest statement
Conflict of Interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
