This is a preprint.
The Spatial Atlas of Human Anatomy (SAHA): A Multimodal Subcellular-Resolution Reference Across Human Organs
- PMID: 40611896
- PMCID: PMC12224548
- DOI: 10.1101/2025.06.16.658716
The Spatial Atlas of Human Anatomy (SAHA): A Multimodal Subcellular-Resolution Reference Across Human Organs
Abstract
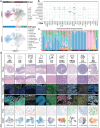
The Spatial Atlas of Human Anatomy (SAHA) represents the first multimodal, subcellular-resolution reference of healthy adult human tissues across multiple organ systems. Integrating spatial transcriptomics, proteomics, and histological features across over 15 million cells from more than 100 donors, SAHA maps conserved and organ-specific cellular niches in gastrointestinal and immune tissues. High-resolution profiling using CosMx SMI, 10x Xenium, RNAscope, GeoMx DSP, and single-nucleus RNA-seq reveals spatially organized cell states, rare adaptive immune populations, and tissue-specific cell-cell interactions. Comparative analyses with colorectal cancer and inflammatory bowel disease demonstrate the power of SAHA to detect disease-associated spatial disruptions, including crypt dedifferentiation, perineural invasion, and therapy-resistant immune remodeling. All data are openly accessible through a FAIR-compliant interactive portal to support exploration, benchmarking, and machine learning model training. Through SAHA, we provide a foundational framework for spatial diagnostics and next-generation precision medicine grounded in a comprehensive human tissue atlas.
Figures

















References
-
- Palla G., Fischer D. S., Regev A. & Theis F. J. Spatial components of molecular tissue biology. Nat. Biotechnol. 40, 308–318 (2022). - PubMed
-
- Rood J. E. et al. The Human Cell Atlas from a cell census to a unified foundation model. Nature 637, 1065–1071 (2025). - PubMed
-
- Velten B. & Stegle O. Principles and challenges of modeling temporal and spatial omics data. Nat. Methods 20, 1462–1474 (2023). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
