An integrative approach prioritizes the orphan GPR61 genomic region in tissue-specific regulation of chronotype
- PMID: 40612388
- PMCID: PMC12224261
- DOI: 10.1093/sleepadvances/zpaf030
An integrative approach prioritizes the orphan GPR61 genomic region in tissue-specific regulation of chronotype
Abstract
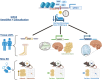
Study objectives: Chronotype, a manifestation of circadian rhythms, affects morning or evening preferences and ease of getting up. This study explores the genetic basis of morning chronotype and ease of getting up, focusing on the G-protein-coupled receptor locus, GPR61.
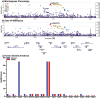
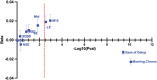
Methods: We analyzed the genetic correlation between chronotype and ease of getting up using linkage disequilibrium score regression with summary statistics from the UK Biobank (n = 453,379). We prioritized shared signals between chronotype and ease of getting up using the Human Genetic Evidence (HuGE) score. We assessed the significance of GPR61 and the lead variant rs12044778 through co-localization and in silico analyses from ENCODE, Genotype-Tissue Expression, Hi-C, and Knockout Mouse Project databases to explore potential regulatory roles of causal genes.
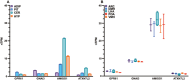
Results: We identified a strong genetic correlation (Rg = 0.80, p = 4.9 × 10324) between chronotype and ease of getting up. Twenty-three genes, including three circadian core clock components, had high HuGE scores for both traits. Lead variant rs12044778 in GPR61 was prioritized for its high HuGE score (45) and causal pleiotropy (posterior probability = 0.98). This morningness variant influenced gene expression in key tissues: decreasing GPR61 in tibial nerve, increasing AMIGO1 in subcutaneous adipose, and increasing ATXN7L2 in the cerebellum. Functional knockout models showed GPR61 knockout increased fat mass and activity, AMIGO1 knockout increased activity, and ATXN7L2 knockout reduced body weight without affecting activity.
Conclusions: Our findings reveal pleiotropic genetic factors influencing chronotype and ease of getting up, emphasizing GPR61's rs12044778 and nearby genes like AMIGO1 and ATXN7L2. These insights advance our understanding of circadian preferences and suggest potential therapeutic interventions.
Keywords: AMIGO1; ATXN7L2; GPR61; circadian rhythm; ease of getting up; genetic pleiotropy; knockout models; morning chronotype; rs12044778.
© The Author(s) 2025. Published by Oxford University Press on behalf of Sleep Research Society.
Figures



Update of
-
An integrative approach prioritizes the orphan GPR61 genomic region in tissue-specific regulation of chronotype.bioRxiv [Preprint]. 2024 Nov 25:2024.11.22.624721. doi: 10.1101/2024.11.22.624721. bioRxiv. 2024. Update in: Sleep Adv. 2025 May 18;6(2):zpaf030. doi: 10.1093/sleepadvances/zpaf030. PMID: 39651283 Free PMC article. Updated. Preprint.
References
-
- Jones SE, Lane JM, Wood AR, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):343. doi: https://doi.org/ 10.1038/s41467-018-08259-7 - DOI - PMC - PubMed
-
- Adan A, Archer SN, Hidalgo MP, Di Milia L, Natale V, Randler C.. Circadian typology: a comprehensive review. Chronobiol Int. 2012;29(9):1153–1175. doi: https://doi.org/ 10.3109/07420528.2012.719971 - DOI - PubMed
-
- Schmid SM, Hallschmid M, Schultes B.. The metabolic burden of sleep loss. Lancet Diabetes Endocrinol. 2015;3(1):52–62. doi: https://doi.org/ 10.1016/S2213-8587(14)70012-9 - DOI - PubMed
-
- Takahashi JS, Hong HK, Ko CH, McDearmon EL.. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: https://doi.org/ 10.1038/nrg2430 - DOI - PMC - PubMed
-
- Jansen PR, Watanabe K, Stringer S, et al. ; 23andMe Research Team. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. doi: https://doi.org/ 10.1038/s41588-018-0333-3 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

