Coupling of glucose metabolism with mitophagy via O-GlcNAcylation of PINK1
- PMID: 40612683
- PMCID: PMC12224001
- DOI: 10.7150/ijbs.112672
Coupling of glucose metabolism with mitophagy via O-GlcNAcylation of PINK1
Abstract
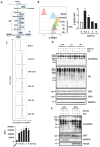
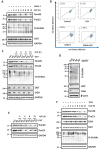
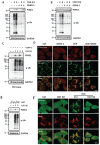
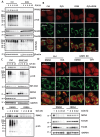
Mitophagy is a selective form of autophagy for the clearance of damaged and dysfunctional mitochondria via the autophagy-lysosome pathway. As mitochondria are the most important metabolic organelles, the process of mitophagy is tightly regulated by glucose metabolism. At present, it is known that glucose is required for the mitophagy process, while the underlying mechanisms remain to be further elucidated. In this study, we establish a novel regulatory role of glucose metabolism in mitophagy via protein O-GlcNAcylation. First, we found that acute mitochondrial damage enhanced glucose uptake and promoted protein O-GlcNAcylation. Second, we provided evidence that protein O-GlcNAcylation promotes PINK1-Parkin-dependent mitophagy. Next, we attempted to illustrate the molecular mechanisms underlying the regulation of O-GlcNAcylation in mitophagy by focusing on PTEN-induced kinase 1 (PINK1). One important observation is that PINK1 is O-GlcNAcylated upon acute mitochondrial damage, and suppression of O-GlcNAcylation impairs PINK1 protein stability and its phosphorylated ubiquitin, leading to impaired mitophagy. More importantly, we found that glucose metabolism promotes mitophagy via regulating O-GlcNAcylation. Taken together, this study demonstrates a novel regulatory mechanism connecting glucose metabolism with mitophagy via O-GlcNAcylation of PINK1. Therefore, targeting the O-GlcNAcylation may provide new strategies for the modulation of mitophagy and mitophagy-related human diseases.
Keywords: HBP; O-GlcNAcylation; PINK1; glucose metabolism; mitophagy.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

