Developing a prognostic signature and characterizing the tumor microenvironment based on centrosome-related genes in lung adenocarcinoma
- PMID: 40612866
- PMCID: PMC12215551
- DOI: 10.32604/or.2025.056176
Developing a prognostic signature and characterizing the tumor microenvironment based on centrosome-related genes in lung adenocarcinoma
Abstract
Background: The centrosome, a crucial cellular structure involved in the mitotic process of eukaryotic cells, plays a significant role in tumor progression by regulating the growth and differentiation of neoplastic cells. This makes the centrosome a promising target for therapeutic strategies in cancer treatment.
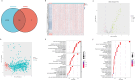
Methods: Utilizing data from the TCGA database, we identified centrosome-related genes and constructed a prognostic model for 518 lung adenocarcinoma patients. Prognosis-associated genes were initially screened using univariate Cox regression, with overfitting minimized by applying LASSO regression to remove collinearity. Finally, a set of 12 genes was selected through multivariable Cox regression for inclusion in the prognostic model.
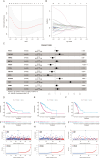
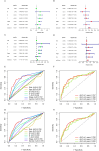
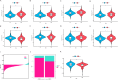
Results: The model's performance was assessed using ROC curve analysis, demonstrating a robust predictive ability with an AUC of 0.728 in the training group and 0.695 in the validation group. Differential expression analysis between high-risk (HRLAs) and low-risk (LRLAs) individuals was performed, followed by enrichment analyses using KEGG, GO, Progeny, GSVA, and GSEA. These analyses revealed significant differences in immune-related pathways between the two groups. Immune microenvironment assessment through ssGSEA and ESTIMATE indicated that individuals with poor prognosis exhibited lower immune, stromal, and ESTIMATE scores, along with higher tumor purity, suggesting an impaired immune microenvironment in HRLAs patients. Drug susceptibility analysis and molecular docking showed that HRLAs individuals were more responsive to docetaxel, emphasizing the therapeutic relevance of paclitaxel in this cohort.
Conclusion: We successfully developed and validated a centrosome-associated gene-based prognostic model, offering clinicians valuable insights for improved decision-making and personalized treatment strategies. This model may facilitate the identification of high-risk patients and guide therapeutic interventions in lung adenocarcinoma.
Keywords: Centrosome; Lung adenocarcinoma; Neoplastic microenvironment; Personalized treatment; Prognostic model.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest to report regarding the present study.
Figures










References
-
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. doi: 10.1097/JTO.0b013e318206a221; - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
