Multiomic integration reveals subtype-specific predictors of neoadjuvant treatment response in breast cancer
- PMID: 40614190
- PMCID: PMC12227054
- DOI: 10.1126/sciadv.adu1521
Multiomic integration reveals subtype-specific predictors of neoadjuvant treatment response in breast cancer
Abstract
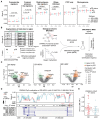
Neoadjuvant therapy has been widely used in breast cancer, but treatment response varies among individuals. We conducted multiomic profiling on tumor samples from 149 Chinese patients with breast cancer across ER-HER2+, ER+HER2+, and ER-HER2- subtypes, categorizing outcomes as pathologic complete response (pCR; n = 81) or residual disease (RD; n = 68). We identified distinct molecular features linked to pCR in each subtype: elevated cell proliferation in patients with ER-HER2- pCR, higher CDKN2A methylation in patients with ER-HER2- RD, increased KIT methylation in patients with ER-HER2+ RD, and MAP4K1 hypermethylation in patients with ER+HER2+ RD. These findings were subsequently validated in independent datasets. By integrating clinical and multiomic data, we developed MOPCR, a subtype-specific machine learning model that outperformed single-omic approaches in predicting treatment response. MOPCR demonstrated potential generalizability across cohorts and provided preliminary stratification of patient subgroups with higher pCR probability, offering valuable insights for precision cancer management.
Figures





References
-
- Fan L., Strasser-Weippl K., Li J.-J., St Louis J., Finkelstein D. M., Yu K.-D., Chen W.-Q., Shao Z.-M., Goss P. E., Breast cancer in China. Lancet Oncol. 15, e279–e289 (2014). - PubMed
-
- von Minckwitz G., Huang C.-S., Mano M. S., Loibl S., Mamounas E. P., Untch M., Wolmark N., Rastogi P., Schneeweiss A., Redondo A., Fischer H. H., Jacot W., Conlin A. K., Arce-Salinas C., Wapnir I. L., Jackisch C., DiGiovanna M. P., Fasching P. A., Crown J. P., Wülfing P., Shao Z., Caremoli E. R., Wu H., Lam L. H., Tesarowski D., Smitt M., Douthwaite H., Singel S. M., Geyer C. E. Jr., KATHERINE Investigators , Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 380, 617–628 (2019). - PubMed
-
- Spring L. M., Fell G., Arfe A., Sharma C., Greenup R., Reynolds K. L., Smith B. L., Alexander B., Moy B., Isakoff S. J., Parmigiani G., Trippa L., Bardia A., Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: A comprehensive meta-analysis. Clin. Cancer Res. 26, 2838–2848 (2020). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

