Metagenomic data insights into chicken microbiome diversity across various regions of Kazakhstan
- PMID: 40614652
- PMCID: PMC12271053
- DOI: 10.1016/j.psj.2025.105488
Metagenomic data insights into chicken microbiome diversity across various regions of Kazakhstan
Abstract
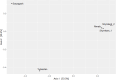
Understanding the gut microbiome of poultry is essential for ensuring the health, productivity, and safety of poultry products. This study aimed to assess the regional diversity and composition of chicken microbiota in Kazakhstan using high-throughput metagenomic sequencing. Tracheal and cloacal swabs were collected from chickens on private farms in five geographic locations. Pooled DNA and RNA samples were sequenced using the Ion Torrent PGM platform, and taxonomic classification was performed using Kaiju, with subsequent alpha and beta diversity analyses in R. The results revealed considerable differences in the microbial profiles between regions. Notably, Chlamydia was abundant in the Shymkent samples (>48 %) but was nearly absent elsewhere. In contrast, Pseudomonas was disproportionately dominant in Almaty (32.7 %), suggesting possible dysbiosis. This study provides the first metagenomic characterization of poultry microbiota in Kazakhstan. This highlights region-specific microbial risks and underscores the importance of spatial microbiome monitoring in poultry health management. These findings provide a basis for future strategies aimed at preventing disease outbreaks and controlling zoonotic pathogens in poultry.
Keywords: Microbiota; Next-generation sequencing; Poultry.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Disclosures The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Ilya Korotetskiy reports financial support was provided by Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Arkin A.P., Cottingham R..W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S., Sneddon M.W., Henderson M.L., Riehl W.J., Murphy-Olson D., Chan S.Y., Kamimura R.T., Kumari S., Drake M.M., Brettin T.S., Glass E.M., Chivian D., Gunter D., Weston D.J., Allen B.H., Baumohl J., Best A.A., Bowen B., Brenner S.E., Bun C.C., Chandonia J.-M., Chia J.-M., Colasanti R., Conrad N., Davis J.J., Davison B.H., DeJongh M., Devoid S., Dietrich E., Dubchak I., Edirisinghe J.N., Fang G., Faria J.P., Frybarger P.M., Gerlach W., Gerstein M., Greiner A., Gurtowski J., Haun H.L., He F., Jain R., Joachimiak M.P., Keegan K.P., Kondo S., Kumar V., Land M.L., Meyer F., Mills M., Novichkov P.S., Oh T., Olsen G.J., Olson R., Parrello B., Pasternak S., Pearson E., Poon S.S., Price G.A., Ramakrishnan S., Ranjan P., Ronald P.C., Schatz M.I.C., Seaver S.M.D., Shukla M., Sutormin R.A., Syed M.H., Thomason J., Tintle N.L., Wang D., Xia F., Yoo H., Yoo S., Yu D. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. - DOI - PMC - PubMed
-
- Wettere A.R.J.V. Avian chlamydiosis. Merck Veterinary Manual. 2024 https://www.merckvetmanual.com/poultry/avian-chlamydiosis/avian-chlamydi...
-
- Lee S.I., Kim S..A., Park S.H., Ricke S.C. In: Food Safety in Poultry Meat Production. Food Microbiology and Food Safety. Venkitanarayanan K., Thakur S., Ricke S., editors. Springer Nature; Switzerland AG: 2019. Molecular and new-generation techniques for rapid detection of foodborne pathogens and characterization of microbial communities in poultry meat; pp. 235–260. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

