Modeling Pseudomonas aeruginosa-Staphylococcus aureus interactions in zebrafish to assess the host inflammatory response upon co-infection
- PMID: 40615487
- PMCID: PMC12227746
- DOI: 10.1038/s41598-025-08459-4
Modeling Pseudomonas aeruginosa-Staphylococcus aureus interactions in zebrafish to assess the host inflammatory response upon co-infection
Abstract
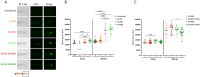
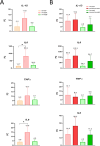
Investigating the interplay between polymicrobial colonization and host response in the context of chronic infections is a complex issue. The interaction between Staphylococcus aureus and Pseudomonas aeruginosa, which frequently co-infect patients with chronic diseases like cystic fibrosis (CF), can be either competitive or coexistent. How these interaction states can influence the host response remains an open question for which relevant infection models are needed. In this study, we investigated co-infections by P. aeruginosa and S. aureus in zebrafish (Danio rerio), a non-mammalian model well established for the study of the host innate immune response. Using a wound infection method and a trio of strains co-isolated from a CF patient, we provide an in vivo co-infection model that recapitulates the competitive and coexistent bacterial interactions observed in vitro. The inflammatory response, monitored through the expression of specific cytokines at the infection site or in the whole larvae, was stronger with P. aeruginosa than S. aureus in the context of mono-infection. Upon co-infection, a competitive or coexistent interaction between P. aeruginosa and S. aureus appeared to slightly modulate this response. Moreover, the RT-qPCR profile of cytokine response observed in zebrafish larvae was similar to the one observed after infection of human lung epithelial cells. Thus, the zebrafish embryo appears as a relevant model to study persistent co-infection with P. aeruginosa and S. aureus, offering unique opportunities to address the host response in this polymicrobial context.
Keywords: Pseudomonas aeruginosa; Staphylococcus aureus; Co-infection; Inflammatory response; Zebrafish model.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Serra, R. et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti-Infe. 13, 605–613 (2015). - PubMed
-
- Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis.18, 318–327 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

