Multi-omics analysis identifies SNP-associated immune-related signatures by integrating Mendelian randomization and machine learning in hepatocellular carcinoma
- PMID: 40615535
- PMCID: PMC12227606
- DOI: 10.1038/s41598-025-09010-1
Multi-omics analysis identifies SNP-associated immune-related signatures by integrating Mendelian randomization and machine learning in hepatocellular carcinoma
Abstract
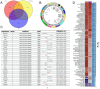
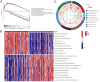
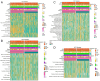
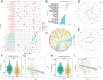
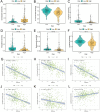
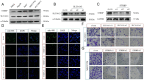
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related death globally, characterized by high morbidity and poor prognosis. The complex molecular and immune landscape of HCC makes accurate patient stratification and personalized treatment essential. In this study, we utilized large-scale gene expression data from TCGA and GSE54236, alongside eQTL GWAS data, to identify key genes that influence HCC prognosis. Machine learning analysis was performed on the genes identified through Mendelian randomization (MR) and survival association analysis, using 101 algorithms to construct a robust prognostic model. A novel riskScore model was developed by integrating genetic, clinical, and immune cell infiltration data. The prognostic performance of model was validated through survival analysis, and its association with chemotherapy and immunotherapy sensitivity. The impact of key genes on the proliferation and invasion capabilities of HCC cells was assessed through Western blot (WB), EdU, and invasion assays. A total of 27 candidate genes associated with HCC survival were identified, with 16 genes categorized as high-risk. The riskScore model demonstrated excellent performance in stratifying patients into high-risk and low-risk groups, with C-index exceeding 0.7 for both TCGA and GSE54236 datasets. High-risk patients exhibited poorer prognosis and higher immune cell infiltration, particularly T cells and neutrophils. The model also predicted drug sensitivity, with high-risk patients showing greater sensitivity to chemotherapy agents like 5-Fluorouracil and Paclitaxel. Mutation analysis revealed that TP53 and MUC16 mutations were prevalent in high-risk groups, highlighting their role in HCC progression and therapeutic response. And the key gene SLC16A3 and STRBP can significantly promote the proliferation and invasion ability of HCC cells. Our riskScore model, integrating genetic and immune factors, provides a robust prognostic tool with potential clinical application in patient stratification and chemotherapy decision-making for HCC patients.
Keywords: Chemotherapy sensitivity; Genetic mutation; HCC; Immune checkpoint; Immune microenvironment; MR; Prognostic; RiskScore.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Consent for publication: All authors informed and consent.
Figures




References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71 (3), 209–249 (2021). - PubMed
-
- Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.68 (6), 394–424 (2018). - PubMed
-
- Hou, Z. et al. Use of chemotherapy to treat hepatocellular carcinoma. Biosci. Trends. 16 (1), 31–45 (2022). - PubMed
-
- Llovet, J. M. et al. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol.19 (3), 151–172 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

