Tracing the evolutionary pathway of SARS-CoV-2 through RNA sequencing analysis
- PMID: 40615624
- PMCID: PMC12227573
- DOI: 10.1038/s41598-025-09911-1
Tracing the evolutionary pathway of SARS-CoV-2 through RNA sequencing analysis
Abstract
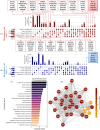
The COVID-19 pandemic, driven by the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), has underscored the need to understand the virus's evolution due to its global health impact. This study employed RNA sequencing (RNA-Seq) to analyze gene expression differences across multiple SARS-CoV-2 variants. We used publicly available datasets from the Gene Expression Omnibus (GEO) with IDs GSE157103, GSE171110, GSE189039, and GSE201530, which contain RNA-Seq data extracted from white blood cells, whole blood, or PBMCs of individuals infected with the Original Wuhan variant (both hospitalized and non-hospitalized), the French variant (hospitalized), the Beta variant (hospitalized), and the Omicron variant (moderate and mild cases), along with COVID-negative controls. Our first objective was to examine differences in gene expression dynamics using Generalized Linear Models with Quasi-Likelihood F-tests and the Magnitude-Altitude Scoring (GLMQL-MAS) technique, followed by Gene Ontology (GO) and pathway analyses. Our second objective was to employ Cross-MAS to identify a robust set of genes indicative of SARS-CoV-2 infection regardless of the variant and to assess their classification performance. GO and pathway analyses revealed a significant evolutionary shift in how SARS-CoV-2 interacts with the host. Early variants such as the Original Wuhan and French cases primarily affected pathways related to viral replication, including Eukaryotic Translation Elongation and Viral mRNA Translation. In contrast, later variants like Beta and Omicron showed a strategic shift toward modulating and evading the host immune response, engaging immune-related pathways such as Interferon Alpha/Beta signaling and Cytokine signaling in the immune system. To evaluate the classification potential of the identified genes, we tested them on held-out datasets GSE152418, PMC8202013, GSE161731, and GSE166190, which contain RNA-Seq data from whole blood or PBMCs of COVID-positive and healthy individuals. Using top-ranked genes such as IFI27, CDC20, RRM2, HJURP, and CDC45 in linear models including logistic regression and linear SVM, we achieved 97.31% accuracy, with precision and recall rates of 0.97 and 0.99, respectively. These signatures also achieved perfect classification (100% accuracy, precision, and recall) in two additional datasets: GSE294888, which includes blood-derived plasmacytoid dendritic cells (pDCs) and type 2 conventional dendritic cells (DC2s) stimulated with Delta or Omicron variants, and GSE239595, which features Omicron-infected nasopharyngeal tissue. These findings demonstrate the potential of transcriptomic signatures for variant-agnostic COVID-19 detection and provide a foundation for flexible diagnostic and therapeutic approaches in response to SARS-CoV-2 evolution.
Keywords: Diagnostic biomarkers; Gene Ontology (GO); Gene expression analysis; Machine learning; Pathway analysis; RNA sequencing (RNA-Seq); SARS-CoV-2 variants.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures







References
-
- Ciotti, M. et al. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci.57(6), 365–388 (2020). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

