The synthetic lethal interaction between CDS1 and CDS2 is a vulnerability in uveal melanoma and across multiple tumor types
- PMID: 40615675
- PMCID: PMC12283370
- DOI: 10.1038/s41588-025-02222-1
The synthetic lethal interaction between CDS1 and CDS2 is a vulnerability in uveal melanoma and across multiple tumor types
Abstract
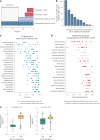
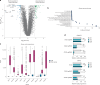
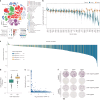
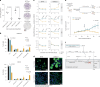
Metastatic uveal melanoma is an aggressive disease with limited effective therapeutic options. To comprehensively map monogenic and digenic dependencies, we performed CRISPR-Cas9 screening in ten extensively profiled human uveal melanoma cell line models. Analysis involved genome-wide single-gene and combinatorial paired-gene CRISPR libraries. Among our 76 uveal melanoma-specific essential genes and 105 synthetic lethal gene pairs, we identified and validated the CDP-diacylglycerol synthase 2 gene (CDS2) as a genetic dependency in the context of low CDP-diacylglycerol synthase 1 gene (CDS1) expression. We further demonstrate that CDS1/CDS2 forms a synthetic lethal interaction in vivo and reveal that CDS2 knockout results in the disruption of phosphoinositide synthesis and increased cellular apoptosis and that re-expression of CDS1 rescues this cell fitness defect. We extend our analysis using pan-cancer data, confirming increased CDS2 essentiality in diverse tumor types with low CDS1 expression. Thus, the CDS1/CDS2 axis is a therapeutic target across a range of cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.J.A. has received precompetitive funding from AstraZeneca and OpenTargets. F.I. provides consultancy services for the joint Cancer Research Horizons–AstraZeneca Functional Genomics Centre and for Mosaic Therapeutics. F.I. also receives funding from OpenTargets and Nerviano Medical Sciences Srl. The remaining authors declare no competing interests.
Figures






References
-
- Khoja, L. et al. Meta-analysis in metastatic uveal melanoma to determine progression free and overall survival benchmarks: an international rare cancers initiative (IRCI) ocular melanoma study. Ann. Oncol.30, 1370–1380 (2019). - PubMed
-
- Nathan, P. et al. Overall survival benefit with tebentafusp in metastatic uveal melanoma. N. Engl. J. Med.385, 1196–1206 (2021). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

