Brain macrophages and pial fibroblasts promote inflammation in a hypomyelination model
- PMID: 40615895
- PMCID: PMC12226894
- DOI: 10.1186/s40478-025-02063-3
Brain macrophages and pial fibroblasts promote inflammation in a hypomyelination model
Abstract
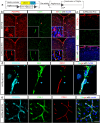
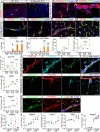
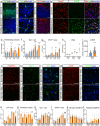
Many neurological diseases remain difficult to treat, necessitating further elucidation of their pathogenesis. Conditional inactivation of Pdgfra in Nestin-expressing cells leads to the depletion of platelet-derived growth factor receptor-alpha+ (PDGFRα+) oligodendroglial lineage cells responsible for myelination, resulting in forebrain hypomyelination and severe, progressive neurological deficits in neonatal mice. The present study examined the cerebral cortex of these mice to better understand the mechanisms underlying such progressive neurological deficits, that are often observed in refractory neurological diseases. Histological and single-cell RNA sequencing analyses showed that, following activation of meningeal border-associated macrophages (BAMs), PDGFRα+ fibroblasts that escaped gene inactivation were extensively recruited from the meninges into the hypomyelinated subpial cerebral cortex. Transcriptional reprogramming suggested that these fibroblasts originated from the pial fibroblast lineage and adopted a myofibroblast-like transcriptional phenotype. The recruited fibroblasts established stable cell-cell interactions with activated brain macrophages, including BAMs and microglia, accompanied by signaling pathways associated with chronic, tissue-damaging inflammation. Subsequently, inflammatory cortical lesions emerged, characterized by glial activation, angiogenesis, and neuronal oxidative stress. Treatment with a PDGFRα-neutralizing antibody significantly reduced fibroblast recruitment and mitigated glial activation and angiogenesis. These findings suggest that meningeal BAMs and pial fibroblasts are key contributors to the formation of tissue-damaging subpial cortical lesions. The interactions between brain macrophages and pial fibroblasts may contribute to the mechanisms underlying chronic and progressive neurological deficits and represent potential therapeutic targets for refractory neurological diseases.
Keywords: Angiogenesis; Border-associated macrophage; Fibrosis; Myelination failure; PDGFRα; Perivascular fibroblast.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal procedures were conducted in accordance with guidelines laid out by the Institutional Animal Care and Use Committee at the University of Toyama (University of Toyama, Sugitani, Toyama City, Japan). All study protocols were approved by the Ethics Committee of the University of Toyama. Consent for publication: All authors agree with publication. Competing interests: The authors declare no competing interests.
Figures




References
-
- Armulik A, Genove G, Betsholtz C (2011) Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21:193–215. 10.1016/j.devcel.2011.07.001 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

