Uncovering key markers and therapeutic targets for renal fibrosis in diabetic kidney disease through bulk and single-cell RNA sequencing
- PMID: 40615901
- PMCID: PMC12228302
- DOI: 10.1186/s12967-025-06554-8
Uncovering key markers and therapeutic targets for renal fibrosis in diabetic kidney disease through bulk and single-cell RNA sequencing
Abstract
Background: Diabetic kidney disease (DKD) is the major cause of chronic kidney failure, with tubulointerstitial fibrosis playing a crucial role in disease development. Identifying fibrosis-related genes is crucial for improving diagnosis and developing novel therapies due to the necessity for early detection and effective treatments.
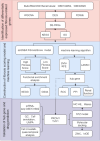
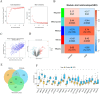
Methods: Genes associated with fibrosis were identified by WGCNA, and a FibrosisScore model was constructed based on ssGSEA scores from two DKD datasets. Essential genes were subsequently confirmed by machine learning and single-cell RNA sequencing (scRNA-seq). Potential therapeutic compounds were identified by screening the ZINC database and confirmed via molecular docking. Critical genes involved in renal fibrosis were analyzed in a streptozotocin (STZ)-induced mouse model of DKD, alongside clinical data from the Nephroseq V5 database.
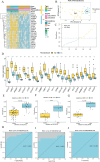
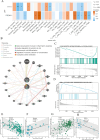
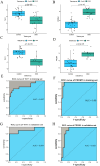
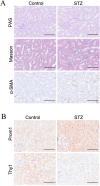
Results: The FibrosisScore model exhibited strong predictive accuracy in both training and validation datasets (AUCs: 0.803, 0.992, 0.891). Patients classified as high-risk demonstrated an increase in M2 macrophages, whereas those identified as low-risk presented a higher prevalence of pro-inflammatory cells. PROM1 and THY1 were recognized as key genes associated with fibrosis. Single-cell RNA analysis revealed that PROM1 is predominantly expressed in proximal tubule cells, while THY1 is enriched in fibroblasts, indicating their distinct roles in fibrosis progression, with both genes exhibiting high diagnostic accuracy (AUC > 0.9). Immune infiltration analysis of PROM1 was primarily associated with a pro-fibrotic, immunosuppressive environment, while THY1 demonstrated antifibrotic properties. ZINC402830 and ZINC3830400 were screened from the ZINC database and validated through molecular docking. In the STZ mouse model, PROM1 correlated with fibrosis and diminished renal function, whereas THY1 exhibited protective effects.
Conclusion: PROM1 and THY1 were critical diagnostic biomarkers for renal fibrosis in DKD, with PROM1 promoting kidney fibrosis and THY1 providing protective effects. The FibrosisScore model demonstrated robust predictive performance, and molecular docking revealed potential therapeutic modulators for these targets.
Keywords: Biomarkers; Diabetic kidney disease; Molecular docking; PROM1; Renal fibrosis; STZ model; Single-cell RNA sequencing; THY1.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The experiments conducted in this research were reviewed and authorized by the Ethics Committee of Zhongnan Hospital at Wuhan University (Ethical Clearance: ZN2024016). Competing interests: None.
Figures





References
-
- Forbes JM, Thorburn DR. Mitochondrial dysfunction in diabetic kidney disease. Nat Rev Nephrol. 2018;14(5):291–312. 10.1038/nrneph.2018.9. - PubMed
-
- van Raalte DH, Bjornstad P, Cherney DZI, de Boer IH, Fioretto P, Gordin D, et al. Combination therapy for kidney disease in people with diabetes mellitus. Nat Rev Nephrol. 2024;20(7):433–46. 10.1038/s41581-024-00827-z. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

