Proteomic Landscape of Colorectal Cancer Derived Liver Metastasis Reveals Three Distinct Phenotypes With Specific Signaling and Enhanced Survival
- PMID: 40617497
- PMCID: PMC12335997
- DOI: 10.1016/j.mcpro.2025.101026
Proteomic Landscape of Colorectal Cancer Derived Liver Metastasis Reveals Three Distinct Phenotypes With Specific Signaling and Enhanced Survival
Abstract
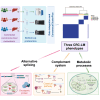
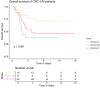
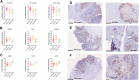
Colorectal carcinoma is a major global disease with the second highest mortality rate among carcinomas. The liver is the most common site for metastases which portends a poor prognosis. Nonetheless, considerable heterogeneity of colorectal cancer liver metastases (CRC-LM) exists, evidenced by varied recurrence and survival patterns in patients undergoing curative-intent resection. Our understanding of the basis for this biological heterogeneity is limited. We investigated this by proteomic analysis of 152 CRC-LM obtained from three different medical centers in Germany and Australia using mass spectrometry-based differential quantitative proteomics. The proteomics data of the individual cohorts were harmonized through batch-effect correction algorithms to build a large multicenter cohort. Applying ConsensusClusterPlus to the proteome data yielded three distinct CRC-LM phenotypes (referred to as CRLM-SD (splice-driven), CRLM-CA (complement-associated), and CRLM-OM (oxidative metabolic)). The CRLM-SD phenotype showed higher abundance of key regulators of alternative splicing as well as extracellular matrix proteins commonly associated with tumor cell growth. The CRLM-CA phenotype was characterized by a higher abundance of proteins involved in the classical pathway part of the complement system including the membrane attack complex proteins and those with antithrombotic activity. The CRLM-OM phenotype showed higher abundance of proteins involved in various metabolic pathways including amino acids and fatty acids metabolism, which correlated in the literature with advanced proliferation of metastases and increased recurrence. Patients classified as CRLM-OM had a significantly lower overall survival in comparison to CRLM-CA patients. Finally, we identified a set of prognosis-associated biomarkers for each group including EpCAM, CEACAM1, CEACAM5, and CEACAM6 for CRLM-SD, DCN, TIMP3, and OLFM4 for CRLM-CA and FMO3, CES2 and AGXT for CRLM-OM. In summary, the discovery of three proteomic subgroups associated with distinct signaling pathways and survival of the CRC-LM patients provides a novel classification for risk stratification, prognosis and potentially novel therapeutic targets in CRC-LM.
Keywords: ECM; EpCAM; alternative splicing; colorectal cancer; complement system; liver metastases; prognosis; proteomics; signaling.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures




References
-
- Ferlay J., Colombet M., Soerjomataram I., Parkin D.M., Piñeros M., Znaor A., et al. Cancer statistics for the year 2020: an overview. Int. J. Cancer. 2021;149:778–789. - PubMed
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer. J. Clini. 2021;71:209–249. - PubMed
-
- Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394:1467–1480. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

