Lactose fermenting enteroinvasive Escherichia coli from diarrhoeal cases confers enhanced virulence
- PMID: 40617833
- PMCID: PMC12228690
- DOI: 10.1038/s41598-025-07232-x
Lactose fermenting enteroinvasive Escherichia coli from diarrhoeal cases confers enhanced virulence
Abstract
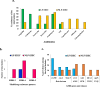
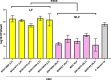
Enteroinvasive Escherichia coli (EIEC), known for causing bacillary dysentery akin to Shigella species, comprises both lactose-fermenting (LF) and non-lactose-fermenting (NLF) isolates. While NLF-EIEC is a well-established pathogen associated with acute dysentery and harbours classical Shigella-like virulence factors, the role of LF-EIEC in human disease remains underexplored. In this study, we sought to characterize LF-EIEC clinical isolates and assessed their pathogenic potential in comparison to NLF-EIEC. Among 13,682 diarrhoeal stool specimens, six LF and nine NLF-EIEC were isolated, predominantly belonging to serogroups O28ac, O125, O136, and O152. Unlike other E. coli, all the EIEC isolates were non-motile. Both the types of EIEC had multiple plasmids harbouring several virulence encoding genes (ipaBCD, ial, virF, sig, sepA and ipaH). Resistance to recent generation antibiotics were mostly confined to NLF-EIEC but some of the LF-EIEC were resistant only to ceftriaxone. Higher invasion ability and significant increase in the expression of virulence encoding genes by the LF-EIEC (p < 0.05) were noted during infection to Int407 cell-line. Additionally, LF-EIEC exhibited extensive colonization of the mouse intestine and expressed severe keratoconjunctivitis in guinea pigs. Together, our findings highlight LF-EIEC as an emerging pathogenic variant warranting heightened surveillance and comprehensive investigation to better understand its epidemiological and clinical significance.
Keywords: Antibiotic resistance; Bacterial infections; Diarrhoea; Enteroinvasive Escherichia coli; Keratoconjunctivitis; Pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The stool samples were collected from patients after obtaining informed written consent from the eligible adult patients or from parents/legal guardians of children. This work was approved by the Institutional Ethical Committee (IEC) of Indian Council of Medical Research- National Institute for Research in Bacterial Infections (ICMR-NIRBI), Kolkata, India (approval no. A-1/2015-IEC). All animal experiments were carried out in accordance with Standard operational procedure of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Ministry of Environment and Forest, Government of India. The protocols for animal studies were approved by Institutional Animal Ethics Committee (IAEC) of ICMR-NIRBI (Registration number: 68/ReBi//S/1999/CPCSEA valid until July 17, 2024). The euthanasia of mice was carried out following the American Veterinary Medical Association (AVMA) guidelines for the Euthanasia of Animals (2020). The animal study was in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. All methods were performed in accordance with the relevant guidelines and regulations and are consistent with the journal policies.
Figures

Similar articles
-
Acquisition of a large virulence plasmid (pINV) promoted temperature-dependent virulence and global dispersal of O96:H19 enteroinvasive Escherichia coli.mBio. 2023 Aug 31;14(4):e0088223. doi: 10.1128/mbio.00882-23. Epub 2023 May 31. mBio. 2023. PMID: 37255304 Free PMC article.
-
Emergence of porcine-derived hybrid Escherichia coli with dual-disease manifestations in intestinal and extraintestinal niches.Virulence. 2025 Dec;16(1):2525927. doi: 10.1080/21505594.2025.2525927. Epub 2025 Jul 2. Virulence. 2025. PMID: 40602993 Free PMC article.
-
Virulence factor profiles, phylogenetic background, and antimicrobial resistance pattern of lactose fermenting and nonlactose fermenting Escherichia coli from extraintestinal sources.Indian J Pathol Microbiol. 2016 Apr-Jun;59(2):180-4. doi: 10.4103/0377-4929.182032. Indian J Pathol Microbiol. 2016. PMID: 27166036
-
Identification and management of Shigella infection in children with diarrhoea: a systematic review and meta-analysis.Lancet Glob Health. 2017 Dec;5(12):e1235-e1248. doi: 10.1016/S2214-109X(17)30392-3. Lancet Glob Health. 2017. PMID: 29132613 Free PMC article.
-
Probiotics for preventing urinary tract infection in people with neuropathic bladder.Cochrane Database Syst Rev. 2017 Sep 8;9(9):CD010723. doi: 10.1002/14651858.CD010723.pub2. Cochrane Database Syst Rev. 2017. PMID: 28884476 Free PMC article.
References
-
- Michelacci, V. et al. Characterization of an emergent clone of enteroinvasive Escherichiacoli circulating in Europe. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis.22(287), e11-19 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

