Somatic loss-of-function mutations in CIDEB reduce hepatic steatosis by increasing lipolysis and fatty acid oxidation
- PMID: 40618957
- PMCID: PMC12313214
- DOI: 10.1016/j.jhep.2025.06.021
Somatic loss-of-function mutations in CIDEB reduce hepatic steatosis by increasing lipolysis and fatty acid oxidation
Abstract
Background & aims: Somatic and germline CIDEB mutations are associated with protection from chronic liver diseases. The mechanistic basis and whether CIDEB suppression would be an effective therapy against fatty liver disease remain unclear.
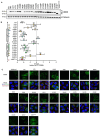
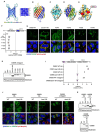
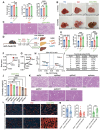
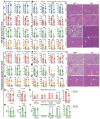
Methods: Twenty-one CIDEB somatic mutations were introduced into cells to assess functionality. In vivo screening was used to trace Cideb mutant clones in mice fed normal chow, western diet (WD), and choline-deficient, L-amino acid-defined, high-fat diet (CDA-HFD) diets. Constitutive and conditional Cideb knockout mice were generated to study Cideb in liver disease. Isotope tracing was used to evaluate fatty acid oxidation and de novo lipogenesis. Transcriptomics, lipidomics, and metabolic analyses were utilized to explore molecular mechanisms. Double knockout models (Cideb/Atgl and Cideb/Pparα) tested mechanisms underlying Cideb loss.
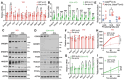
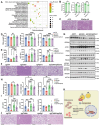
Results: Most CIDEB mutations impaired function, and loss-of-function clones were positively selected under CDA-HFD but not all steatogenic diets. Cideb knockout mice were protected from WD-, CDA-HFD-, and alcohol-induced liver disease, with the strongest effect in CDA-HFD models. Hepatocyte-specific Cideb deletion ameliorated disease after MASLD (metabolic dysfunction-associated steatotic liver disease) establishment, modeling the impact of therapeutic small-interfering RNAs. Cideb loss protected livers via increased β-oxidation, specifically through ATGL and PPARα activation.
Conclusions: Cideb deletion is more protective in some types of fatty liver disease. β-oxidation is an important component of the Cideb protective mechanism. CIDEB inhibition represents a promising approach, and somatic mutations in CIDEB might predict the patient populations who will benefit the most.
Impact and implications: It is not clear why somatic and germline CIDEB mutations are protective in metabolic dysfunction-associated steatotic liver disease (MASLD). Cideb mutations are predominantly loss of function, and Cideb-deficient clones selectively expand in specific dietary contexts such as choline-deficient, L-amino acid-defined, high-fat diet-induced MASLD. Consistently, liver-wide deletion of Cideb ameliorates MASLD most profoundly after choline-deficient, L-amino acid-defined, high-fat diet feeding. Mechanistically, Cideb deficiency enhances hepatic fatty acid β-oxidation via ATGL and PPARα activation. These findings suggest that CIDEB inhibition might be most effective in patients with the subtypes of MASLD that promote the expansion of CIDEB mutant clones.
Keywords: CIDEB; fatty liver disease; in vivo screening; somatic mutations; β-oxidation.
Copyright © 2025 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest H.Z. and P.C. are co-founders of Quotient Therapeutics and Jumble Therapeutics. H.Z. is an advisor for Newlimit, Alnylam Pharmaceuticals, and Chroma Medicines. H.Z. received research support from Chroma Medicines and owns stock in Ionis Pharmaceuticals. H.Z. and L.L. have a patent on CIDEB siRNA for liver disease (patent #63/328,557). Please refer to the accompanying ICMJE disclosure forms for further details.
Figures



References
-
- Ng SWK, Rouhani FJ, Brunner SF, et al. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021;598:473–8. - PubMed
-
- Verweij N, Haas ME, Nielsen JB, et al. Germline Mutations in CIDEB and Protection against Liver Disease. N Engl J Med. 2022;387:332–44. - PubMed
-
- Li JZ, Ye J, Xue B, et al. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 2007;56:2523–32. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

