Exosomes From Intestinal Epithelial Cells Promote Hepatic Differentiation of Liver Progenitor Cells in Gut-Liver-on-a-Chip Models
- PMID: 40619613
- PMCID: PMC12407332
- DOI: 10.1002/advs.202417478
Exosomes From Intestinal Epithelial Cells Promote Hepatic Differentiation of Liver Progenitor Cells in Gut-Liver-on-a-Chip Models
Abstract
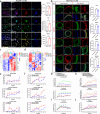
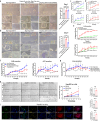
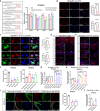
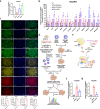
Hepatic progenitor cells (HPCs) are frequently overactivated, and their differentiation into hepatocytes is impaired in advanced liver diseases. To explore the effects of intestinal epithelial cells and their exosomes on the hepatic differentiation of HPCs, co-culture systems of Caco-2/HepaRG cell lines and intestine/HPC organoids are established in a novel gut-liver-on-a-chip. Exosomes derived from intestinal organoids are administered to mice with carbon tetrachloride (CCL4)-induced liver fibrosis. The results showed that the co-culture of HPCs and intestinal epithelial cells promoted the hepatic differentiation of HPCs, mediated by exosomes derived from intestinal epithelial cells. Treatment with exosomes derived from intestinal organoids ameliorated liver fibrosis in a mouse model of CCL4-induced liver fibrosis. A cluster of miRNAs, miR-371-373, is identified within the exosomes of the intestinal epithelial cells, which target RPS6KA2 to modulate hepatic differentiation. This findings demonstrate that exosomes from intestinal epithelial cells promote the hepatic differentiation of HPCs. Exosomes from intestinal organoids may be a novel therapeutic strategy for the treatment of advanced liver diseases.
Keywords: exosomes; hepatic progenitor cells; intestinal epithelial cells; liver fibrosis; organ‐on‐a‐chip.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Xiao J., Wang F., Wong N. K., He J. H., Zhang R., Sun R. J., Xu Y. Y., Liu Y. X., Li W., Koike K. Z., He W. L., You H., Miao Y. L., Liu X. W., Meng M. M., Gao B., Wang H., Li C., J. Hepatol. 2019, 71, 212. - PubMed
-
- Michalopoulos G. K., Bhushan B., Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
