Haploinsufficient variants in SMAD5 are associated with isolated congenital heart disease
- PMID: 40619738
- PMCID: PMC12305712
- DOI: 10.1016/j.xhgg.2025.100478
Haploinsufficient variants in SMAD5 are associated with isolated congenital heart disease
Abstract
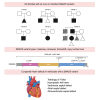
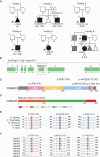
Mothers against decapentaplegic homolog 5 (SMAD5) is a transcriptional regulator that functions within the TGF-β signaling cascade. Evidence from animal studies show that it is crucial for dorsoventral patterning, left-right asymmetry, cardiac looping, and other embryonic processes. However, its role in human development has not been explored, and the contribution of SMAD5 variants to congenital disease is unknown. Here, we report SMAD5 variants identified in six unrelated families with seven individuals presenting with congenital heart disease (CHD). Isolated congenital heart defects are observed in six individuals who carry de novo or inherited missense, nonsense, frameshift, or copy-number variants in SMAD5. A multi-organ phenotype is observed in one individual with a de novo SMAD5 variant that alters an amino acid crucial for SMAD5 multimerization. Septal defects, identified in four individuals, are the most common cardiac lesion in our cohort, with hypoplastic left heart also observed in two individuals. In silico assessment of SMAD5 missense variants predicts disrupted binding to co-factors, and in vitro functional assessment shows changes in SMAD5 gene and protein expression, as well as impaired activation of a BMP4-responsive promoter by the variants. Our findings suggest haploinsufficiency as the underlying molecular mechanism in five of the six families, resulting in isolated CHD, with a SMAD5 dominant-negative variant identified in one family leading to multiple congenital defects. Here, we provide evidence that SMAD5 variants lead to CHD and offer a basis for future exploration of SMAD5 variants in both CHD and post-natal disease.
Keywords: BMP signaling; SMAD5 signaling; TGF-β signaling; cardiac septal defects; congenital heart disease; hypoplastic left heart.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

