Mendelian Randomization Analysis of Mitochondria-Related Genes and Screening of Prognostic Genes in Colorectal Cancer
- PMID: 40620061
- PMCID: PMC12230350
- DOI: 10.1002/cam4.71012
Mendelian Randomization Analysis of Mitochondria-Related Genes and Screening of Prognostic Genes in Colorectal Cancer
Abstract
Background: Mitochondria have been linked with inflammatory colorectal cancer (CRC) development; however, the association between mitochondria-related genes (MRGs) and CRC remains unknown.
Aims: To explore the causal relationship between MRGs and CRC, screen prognostic genes, conduct drug prediction analyses, and investigate the correlations between prognostic genes and immune cells.
Materials and methods: We obtained 1136 MRGs from the MitoCarta3.0 database and analyzed the causal relationship between MRGs expression, methylation, and protein abundance and CRC by Mendelian randomization and sensitivity testing. Prognostic genes were screened via protein-protein interaction networks, enrichment, multi-omics, and survival analyses. Selected key genes were subjected to drug prediction analyses. The prognostic genes and immune cell correlations were explored using Spearman's correlation.
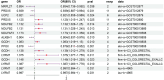
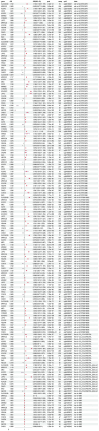
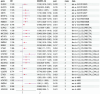
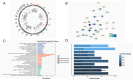
Results: The results indicated that 44 MRGs showed causal relationships with CRC. Six genes (sterol carrier protein2 [SCP2], ATP binding cassette subfamily D member 3 [ABCD3], cytochrome coxidase assembly factor heme A: farnesyltransferase [COX10], mitochondrial contact site and cristae organizing system subunit 10 [MiCOS 10], glutaryl-Coenzyme A dehydrogenase [GCDH], and mitochondrial translational release factor 1-like [MTRF1L] were causally associated with CRC and showed better prognostic significance when their expression levels were high, and there were 106 drugs targeting them. SCP2, ABCD3, MICOS10, GCDH, and MTRF1L were associated with most immune cells, while COX10 was not associated with any of the 96 immune cells.
Discussion: The identification of causal MRGs and their prognostic significance provides new insights into mitochondria's role in CRC. Drug prediction and immune correlations may guide therapy, but validation in larger cohorts and models is needed.
Conclusion: This study reveals causal associations between specific MRGs and CRC, identifies prognostic genes with therapeutic potential, and clarifies immune cell relationships, advancing CRC pathogenesis understanding and treatment development.
Keywords: Mendelian randomization; colorectal cancer; mitochondria‐related genes; potential targets; prognosis.
© 2025 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Potential of SPHK1 as a prognostic marker and therapeutic target in colorectal cancer: insights from bioinformatics and experimental analysis.Int J Surg. 2025 Jul 1;111(7):4550-4575. doi: 10.1097/JS9.0000000000002506. Epub 2025 May 28. Int J Surg. 2025. PMID: 40434736
-
Mito-fission gene prognostic model for colorectal cancer.PeerJ. 2025 Jun 18;13:e19522. doi: 10.7717/peerj.19522. eCollection 2025. PeerJ. 2025. PMID: 40547309 Free PMC article.
-
Identification of mitochondria-related biomarkers in childhood allergic asthma.BMC Med Genomics. 2024 May 23;17(1):141. doi: 10.1186/s12920-024-01901-y. BMC Med Genomics. 2024. PMID: 38783263 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Chemoprevention of colorectal cancer: systematic review and economic evaluation.Health Technol Assess. 2010 Jun;14(32):1-206. doi: 10.3310/hta14320. Health Technol Assess. 2010. PMID: 20594533
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

