The Expression of lncRNAs EVADR and LUESCC in Colorectal Tumor Tissues and Their Association With the CRC Risk
- PMID: 40620190
- PMCID: PMC12230507
- DOI: 10.1002/cnr2.70232
The Expression of lncRNAs EVADR and LUESCC in Colorectal Tumor Tissues and Their Association With the CRC Risk
Abstract
Background: Colorectal cancer (CRC) is a prevalent form of cancer globally and ranks as the second most common cause of cancer-related deaths. Long non-coding RNAs (lncRNAs) are regulatory RNAs that influence gene expression. EVADR and LUESCC are two novel lncRNAs specifically expressed in tumors of glandular origin, such as the colon.
Aims: This study aimed to investigate the expression of EVADR and LUESCC in colorectal tumor tissues and evaluate their potential as diagnostic and prognostic biomarkers in CRC.
Methods: Fifty cases of colorectal tumor tissues, formalin-fixed, paraffin-embedded (FFPE) from individuals with sporadic CRC, referred from the Pathology Department of Imam Hossein Hospital in Tehran, Iran, were analyzed. The expression patterns of LUESCC and EVADR lncRNAs in CRC patients were examined.
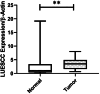
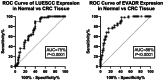
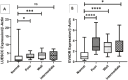
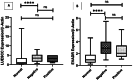
Results: The study reveals upregulation of LUESCC and EVADR lncRNAs in colorectal cancer (CRC) patients compared to normal tissues, with fold changes of 3.52 (p < 0.001) for LUESCC and 3.08 (p < 0.001) for EVADR. ROC curve analysis indicates an area under the curve (AUC) of 0.75 for LUESCC and 0.86 for EVADR, suggesting strong diagnostic potential. Additionally, differential expression analysis shows correlations between lncRNA levels and tumor differentiation grades.
Conclusion: This study highlights the potential of LUESCC and EVADR lncRNAs as biomarkers for CRC diagnosis and prognosis. However, limitations include a small sample size that may affect the generalizability of the findings and a lack of functional assays to elucidate their roles in tumor biology. Further research is needed to validate these findings and explore the underlying mechanisms.
Keywords: EVADR; LUESCC; bioinformatics; colorectal cancer; long non‐coding RNAs.
© 2025 The Author(s). Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Long non-coding RNA SNHG7 as a key driver of colorectal cancer proliferation and metastasis by inhibiting cuproptosis.Eur J Med Res. 2025 Jul 2;30(1):557. doi: 10.1186/s40001-025-02770-6. Eur J Med Res. 2025. PMID: 40604963 Free PMC article.
-
Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer.J Cancer Res Clin Oncol. 2016 Nov;142(11):2291-301. doi: 10.1007/s00432-016-2238-9. Epub 2016 Sep 3. J Cancer Res Clin Oncol. 2016. PMID: 27591862 Free PMC article.
-
Screening and validation of long non-coding RNAs associated with colorectal cancer based on random forest and LASSO regression algorithm.Discov Oncol. 2025 Jul 1;16(1):1217. doi: 10.1007/s12672-025-03048-3. Discov Oncol. 2025. PMID: 40591188 Free PMC article.
-
Role of long non-coding RNAs in neurofibromatosis and Schwannomatosis: pathogenesis and therapeutic potential.Epigenomics. 2024 Dec-Dec;16(23-24):1453-1464. doi: 10.1080/17501911.2024.2430170. Epub 2024 Nov 27. Epigenomics. 2024. PMID: 39601046 Review.
-
Faecal immunochemical tests to triage patients with lower abdominal symptoms for suspected colorectal cancer referrals in primary care: a systematic review and cost-effectiveness analysis.Health Technol Assess. 2017 May;21(33):1-234. doi: 10.3310/hta21330. Health Technol Assess. 2017. PMID: 28643629 Free PMC article.
References
-
- Morgan E., Arnold M., Gini A., et al., “Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimates From GLOBOCAN,” Gut 72, no. 2 (2023): 338–344. - PubMed
-
- Weledji E., “The Etiology and Pathogenesis of Colorectal Cancer,” Clinical Oncology 9 (2024): 2046.
-
- Koley S., “Dr. Tony Dhillon, a British‐Born Medical Oncologist of Indian Origin, the Pioneer of Recent Innovation of Bowel Cancer Vaccine: A Medicometric Portrait,” Journal of Data Science, Informetrics, and Citation Studies 3, no. 2 (2024): 183–205.
-
- World‐Health‐Organization , “Colorectal‐Cancer,” 2024, https://www.who.int/news‐room/fact‐sheets/detail/colorectal‐cancer.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

