Diet and environment drive the convergence of gut microbiome in wild-released giant pandas and forest musk deer
- PMID: 40620903
- PMCID: PMC12226362
- DOI: 10.1016/j.isci.2025.112837
Diet and environment drive the convergence of gut microbiome in wild-released giant pandas and forest musk deer
Abstract
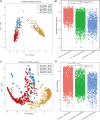
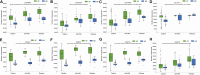
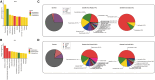
Reintroduction is important for recovering endangered species, and gut microbiome is crucial for successful wildlife reintroduction. This study utilized 16S rRNA high-throughput sequencing of 791 fecal samples to examine the gut microbial changes in giant pandas (Ailuropoda melanoleuca) and forest musk deer (Moschus berezovskii) across captivity, semi-release, and release stages. Our results revealed a similar transitional pattern in the gut microbiome of both species, with semi-release stage displaying an intermediate state between captive and wild microbiome. We also observed that both species are enriched in Pseudomonas and functional pathways related to amino acid metabolism, ATP-binding cassette transporters, and acetyl-CoA/propionyl-CoA carboxylase. Furthermore, the SourceTracker analysis indicated putative contributions of plant and soil microbiome to the gut microbiome of forest musk deer. These findings suggest that similar herbivorous diets and same environment may contribute to the convergence of gut microbiome. In conclusion, our study provides valuable insights for reintroducing endangered wildlife.
Keywords: Microbiology; Nature conservation; Zoology.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources

