Distinguishing PEX2 and PEX16 gene variant severity for mild, severe and atypical peroxisome biogenesis disorders
- PMID: 40621817
- PMCID: PMC12352285
- DOI: 10.1242/dmm.052258
Distinguishing PEX2 and PEX16 gene variant severity for mild, severe and atypical peroxisome biogenesis disorders
Abstract
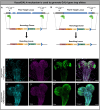
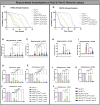
Peroxisomal biogenesis disorders (PBD) are autosomal recessive diseases caused by mutations in specific PEX genes that impair peroxisome formation, leading to multi-systemic failure. Symptoms vary, even in patients with variants in the same PEX gene. Our goal is to select PEX mutations and use Drosophila to model a severity spectrum based on genotype-phenotype correlations. Utilizing KozakGAL4 (KZ) cassettes, we replaced the coding sequence of Pex with a GAL4 driver, ideal for making 'humanized' flies in which human PEX can replace the fly loss. We generated Pex2KZ and Pex16KZ lines and assessed them in various behavior assays, confirming their severe phenotypes. We performed rescue with human reference, variant PEX2 and PEX16 alleles, and phenotypic rescue was observed when human PEX2Ref or PEX16Ref were expressed in Pex2KZ or Pex16KZ flies, respectively. We identified a severity spectrum for PEX2 and PEX16 alleles, with some missense mutations exhibiting severity comparable to truncations. Alleles linked to mild PBD showed partial rescue, while variants associated with atypical ataxia could fully rescue. Drosophila humanization is an effective method to study the range of severity of PBD.
Keywords: Drosophila; PBD; PEX16; PEX2; Peroxisome; Zellweger.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




Update of
-
Distinguishing PEX gene variant severity for mild, severe, and atypical peroxisome biogenesis disorders in Drosophila.bioRxiv [Preprint]. 2024 Nov 19:2024.11.14.623590. doi: 10.1101/2024.11.14.623590. bioRxiv. 2024. Update in: Dis Model Mech. 2025 Jul 1;18(7):dmm052258. doi: 10.1242/dmm.052258. PMID: 39605732 Free PMC article. Updated. Preprint.
References
-
- Bose, M., Yergeau, C., D'Souza, Y., Cuthbertson, D. D., Lopez, M. J., Smolen, A. K. and Braverman, N. E. (2022). Characterization of severity in Zellweger spectrum disorder by clinical findings: a scoping review, meta-analysis and medical chart review. Cells 11, 1891. 10.3390/cells11121891 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

