JAK2 inhibition mediates clonal selection of RAS pathway mutations in myeloproliferative neoplasms
- PMID: 40623967
- PMCID: PMC12234676
- DOI: 10.1038/s41467-025-60884-1
JAK2 inhibition mediates clonal selection of RAS pathway mutations in myeloproliferative neoplasms
Abstract
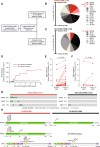
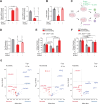
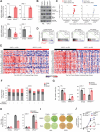
JAK (Janus Kinase) inhibitors, such as ruxolitinib, were introduced a decade ago for treatment of myeloproliferative neoplasms (MPN). To evaluate ruxolitinib's impact on MPN clonal evolution, we interrogate a myelofibrosis patient cohort with longitudinal molecular evaluation and discover that ruxolitinib is associated with clonal outgrowth of RAS pathway mutations. Single-cell DNA sequencing combined with ex vivo treatment of RAS mutated CD34+ primary patient cells, demonstrates that ruxolitinib induces RAS clonal selection both in a JAK/STAT wild-type and hyper-activated context. RAS mutations are associated with decreased transformation-free and overall survival only in patients treated with ruxolitinib. In vitro and in vivo competition assays demonstrate increased cellular fitness of RAS-mutated cells under ruxolitinib or JAK2 knock-down, consistent with an on-target effect. MAPK pathway activation is associated with JAK2 downregulation resulting in enhanced oncogenic potential of RAS mutations. Our results prompt screening for pre-existing RAS mutations in JAK inhibitor treated patients with MPN.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: L.B. received research funding from Gilead and Pfizer for unrelated projects, and personal fees from BMS, Novartis and GSK outside of the submitted work. N.G. received personal fees from Novartis, Abbvie and Astra Zeneca outside of the submitted work. K.S. is on the SAB and has stock options in Auron Therapeutics and received grant funding from Novartis and KronosBio on topics unrelated to this manuscript. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

