Decoding and reprogramming of the biosynthetic networks of mushroom-derived bioactive type II ganoderic acids in yeast
- PMID: 40623979
- PMCID: PMC12234687
- DOI: 10.1038/s41421-025-00812-1
Decoding and reprogramming of the biosynthetic networks of mushroom-derived bioactive type II ganoderic acids in yeast
Abstract
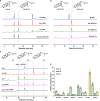
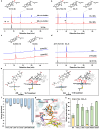
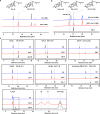
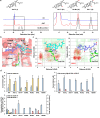
Mushroom's specialized secondary metabolites possess important pharmacological activities, but their biosynthetic pathway elucidation is extremely challenging, not to mention reprogramming of their biosynthetic networks to target metabolites. By taking Ganoderma lucidum, a famous traditional medicinal mushroom, as a lead example, here we decoded the biosynthetic networks of type II ganoderic acids (TIIGAs), a group of its main bioactive metabolites by studying the coordinated gene expression in G. lucidum, identifying endogenous or heterologous enzymes capable of C22 hydroxylation, configuration conversion of C3 hydroxyl group, and acetylation on C3, C15 and C22 hydroxyl groups. Notably, we revealed the catalytic mechanism of the C22 hydroxylase CYP512W6, and an unexpected bifunctional acetyltransferase GlAT that is required to transfer acetyl groups to C15 and C22. Using a fluorescence-guided integration method, we achieved efficient biosynthesis of significant TIIGAs applicable to industrial fermentation. After introducing all the identified enzymes to baker's yeast, we observed that biosynthesis of downstream TIIGAs was severely impeded, and dredged the metabolic block by temporally regulating the expression of acetyltransferases. By reprogramming of the biosynthetic networks of TIIGAs, we were able to produce over 30 TIIGAs, exhibiting 1-4 orders of magnitude higher titers or efficiencies than those from farmed mushrooms. The work enables the access to valuable TIIGAs, facilitates their widespread application, and sheds light on research of other mushroom products.
© 2025. The Author(s).
Conflict of interest statement
Conflict of interest: The enzymes, compounds, and engineered strains described in this study are covered by patents CN2024111285319, CN2024111379443, and CN2024111586867. For the first two patents, H.X., Q.W. and J.J.Z. are listed as co-inventors. For the last patent, H.X., Y.L., Q.W., S.Z., and J.J.Z. are listed as co-inventors.
Figures



Similar articles
-
Ganoderma lucidum (Reishi mushroom) for cancer treatment.Cochrane Database Syst Rev. 2016 Apr 5;4(4):CD007731. doi: 10.1002/14651858.CD007731.pub3. Cochrane Database Syst Rev. 2016. PMID: 27045603 Free PMC article.
-
Sertindole for schizophrenia.Cochrane Database Syst Rev. 2005 Jul 20;2005(3):CD001715. doi: 10.1002/14651858.CD001715.pub2. Cochrane Database Syst Rev. 2005. PMID: 16034864 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Ganoderma lucidum (Reishi mushroom) for cancer treatment.Cochrane Database Syst Rev. 2012 Jun 13;(6):CD007731. doi: 10.1002/14651858.CD007731.pub2. Cochrane Database Syst Rev. 2012. Update in: Cochrane Database Syst Rev. 2016 Apr 05;4:CD007731. doi: 10.1002/14651858.CD007731.pub3. PMID: 22696372 Updated.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
-
- Chang, S. & Buswell, J. Medicinal mushrooms: past, present and future. in Biochemical Engineering and Biotechnology of Medicinal Mushrooms (eds. Berovic, M. & Zhong, J.) 1-27 (Springer International Publishing, Cham, 2023). - PubMed
-
- Sanodiya, B. S., Thakur, G. S., Baghel, R. K., Prasad, G. B. K. S. & Bisen, P. S. Ganoderma lucidum: a potent pharmacological macrofungus. Curr. Pharm. Biotechnol.10, 717–742 (2009). - PubMed
-
- Homer, J. A. & Sperry, J. Mushroom-derived indole alkaloids. J. Nat. Prod.80, 2178–2187 (2017). - PubMed
Grants and funding
- 24HC2810800/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 31971344/National Natural Science Foundation of China (National Science Foundation of China)
- 31770037/National Natural Science Foundation of China (National Science Foundation of China)
- 2060302-2303-02/China Academy of Chinese Medical Sciences (CACMS)
LinkOut - more resources
Full Text Sources
Miscellaneous

