Single-cell transcriptomic analysis identifies systemic immunosuppressive myeloid cells and local monocytes/macrophages as key regulators in polytrauma-induced immune dysregulation
- PMID: 40623986
- PMCID: PMC12234669
- DOI: 10.1038/s41413-025-00444-x
Single-cell transcriptomic analysis identifies systemic immunosuppressive myeloid cells and local monocytes/macrophages as key regulators in polytrauma-induced immune dysregulation
Abstract
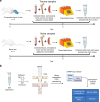
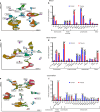
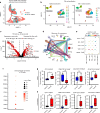
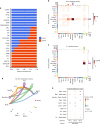
Polytrauma with significant bone and volumetric muscle loss presents substantial clinical challenges. Although immune responses significantly influence fracture healing post-polytrauma, the cellular and molecular underpinnings of polytrauma-induced immune dysregulation require further investigation. While previous studies examined either injury site tissue or systemic tissue (peripheral blood), our study uniquely investigated both systemic and local immune cells at the same time to better understand polytrauma-induced immune dysregulation and associated impaired bone healing. Using single-cell RNA sequencing (scRNA-seq) in a rat polytrauma model, we analyzed blood, bone marrow, and the local defect soft tissue to identify potential cellular and molecular targets involved in immune dysregulation. We identified a trauma-associated immunosuppressive myeloid (TIM) cell population that drives systemic immune dysregulation, immunosuppression, and potentially impaired bone healing. We found CD1d as a global marker for TIM cells in polytrauma. In the local defect tissue, we observed Spp1+ monocytes/macrophages mediating inflammatory, fibrotic, and impaired adaptive immune responses. Finally, our findings highlighted increased signaling via Anxa1-Fpr2 and Spp1-Cd44 axes. This comprehensive analysis enhances our understanding of immune dysregulation-mediated nonunion following traumatic injury and provides biomarkers that could function as treatment targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Ruehle, M. A. et al. Effects of BMP‐2 dose and delivery of microvascular fragments on healing of bone defects with concomitant volumetric muscle loss. J. Orthop. Res.37, 553–561 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

