Nuclear morphometrics coupled with machine learning identifies dynamic states of senescence across age
- PMID: 40624008
- PMCID: PMC12234852
- DOI: 10.1038/s41467-025-60975-z
Nuclear morphometrics coupled with machine learning identifies dynamic states of senescence across age
Abstract
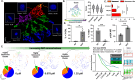
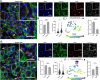
Cellular senescence is an irreversible state of cell cycle arrest with a complex role in tissue repair, aging, and disease. However, inconsistencies in identifying cellular senescence have led to varying conclusions about their functional significance. We developed a machine learning-based approach that uses nuclear morphometrics to identify senescent cells at single-cell resolution. By applying unsupervised clustering and dimensional reduction techniques, we built a robust pipeline that distinguishes senescent cells in cultured systems, freshly isolated cell populations, and tissue sections. Here we show that this method reveals dynamic, age-associated patterns of senescence in regenerating skeletal muscle and osteoarthritic articular cartilage. Our approach offers a broadly applicable strategy to map and quantify senescent cell states in diverse biological contexts, providing a means to readily assess how this cell fate contributes to tissue remodeling and degeneration across lifespan.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: SAM and MNW are listed as inventors on the US Provisional Patent Application #63/671,334, which pertains to the nuclear morphometric pipeline. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

