Conserved missense variant pathogenicity and correlated phenotypes across paralogous genes
- PMID: 40624578
- PMCID: PMC12235991
- DOI: 10.1186/s13059-025-03663-x
Conserved missense variant pathogenicity and correlated phenotypes across paralogous genes
Abstract
Background: The majority of missense variants in clinical genetic tests are classified as variants of uncertain significance. Prior research shows that the deleterious effects and the subsequent molecular consequences of variants are often conserved among paralogous protein sequences within a gene family. Here, we systematically quantify on an exome-wide scale whether the existence of pathogenic variants in paralogous genes at a conserved position can serve as evidence for the pathogenicity of a new variant. For the gene family of voltage-gated sodium channels, where variants and expert-curated clinical phenotypes are available, we also assess whether phenotype patterns of multiple disorders for each gene are conserved across variant positions within the gene family.
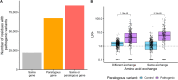
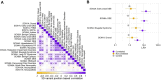
Results: Mapping 590,000 pathogenic and 1.9 million population variants onto 9928 genes grouped into 2054 paralogous families increases the number of residues with classifiable evidence 5.1-fold compared with gene-specific data alone. The presence of a pathogenic variant in a paralogous gene is associated with a positive likelihood ratio of 13.0 for variant pathogenicity. Across ten genes encoding voltage-gated sodium channels and 22 expert-curated disorders, we identify cross-paralog correlated phenotypes based on 3D structure spatial position. For example, multiple established loss-of-function related disorders across SCN1A, SCN2A, SCN5A, and SCN8A show overlapping spatial variant clusters. Finally, we show that phenotype integration in paralog variant selection improves variant classification.
Conclusion: Conserved pathogenic missense variants in paralogous genes provide robust, quantifiable support for clinical variant interpretation, and phenotype-informed mapping further improves predictions.
Keywords: ACMG; Genetics; missense; sodium channel; variant classification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate.: Not applicable. Consent for publication: Not applicable. Competing interests: The authors report no competing interests.
Figures


References
-
- Milman A, et al. Genotype-phenotype correlation of SCN5A genotype in patients with brugada syndrome and arrhythmic events: insights from the SABRUS in 392 probands. Circ Genom Precis Med. 2021;14:e003222. - PubMed
-
- Kamada F, et al. A novel KCNQ4 one-base deletion in a large pedigree with hearing loss: implication for the genotype-phenotype correlation. J Hum Genet. 2006;51:455–60. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

