Meta-Transcriptomes From Microcosms From a Cr Impacted Soil Provides Insights Into the Metabolic Response of the Microbial Populations to Acetate Stimulation
- PMID: 40624790
- PMCID: PMC12234377
- DOI: 10.1111/1758-2229.70148
Meta-Transcriptomes From Microcosms From a Cr Impacted Soil Provides Insights Into the Metabolic Response of the Microbial Populations to Acetate Stimulation
Abstract
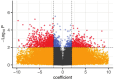
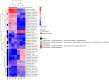
Environmental contamination by Cr(VI) leaching from chromite ore processing residue (COPR) legacy disposal sites can pose a threat to human health. Under iron-reducing conditions, microbial activity can convert mobile and toxic Cr(VI) to less mobile and less toxic Cr(III); however, COPR waste is a very hostile environment for microbial life. Microcosms using soil from beneath a COPR disposal site were challenged with Cr(VI) with and without acetate to stimulate microbial metabolism. Geochemistry showed that when the microbial populations were reducing iron, Cr(VI) was also reduced, and 16S rRNA gene sequencing showed that the community composition evolved over the course of the experiment. Meta-transcriptome data revealed ~3% of transcripts were differentially regulated (p = 0.01) between the acetate amended and unamended systems, with twice as many transcripts downregulated by acetate. Gene ontology (GO) terms for processes involving the cell wall, cell periphery, plasma membrane and encapsulating structures as well as catabolic processes, especially carbohydrate metabolism, were significantly enriched in the unamended microcosm meta-transcriptome. Transcripts for alternative sigma (σ) factors and anti-σ factors were prominent among the differentially regulated genes. The study provides insight into how the provision of acetate shapes metabolic processes and life history strategies in an alkaline Cr(VI) impacted environment.
Keywords: COPR; acetate; alkaline environment; bioremediation; chromium; meta‐transcriptome.
© 2025 The Author(s). Environmental Microbiology Reports published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Metagenomes from microbial populations beneath a chromium waste tip give insight into the mechanism of Cr (VI) reduction.Sci Total Environ. 2024 Jun 25;931:172507. doi: 10.1016/j.scitotenv.2024.172507. Epub 2024 Apr 23. Sci Total Environ. 2024. PMID: 38657818
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Cr(VI)-bioremediation mechanism of a novel strain Bacillus sp. Cr02 with simultaneous aerobic/anaerobic reduction for Cr(VI) from soil to groundwater.J Hazard Mater. 2025 Sep 5;495:139034. doi: 10.1016/j.jhazmat.2025.139034. Epub 2025 Jun 23. J Hazard Mater. 2025. PMID: 40578189
-
Assessing the efficiency and the side effects of atrazine-degrading biocomposites amended to atrazine-contaminated soil.FEMS Microbiol Ecol. 2025 Jun 24;101(7):fiaf071. doi: 10.1093/femsec/fiaf071. FEMS Microbiol Ecol. 2025. PMID: 40580107 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Anderson, R. A. 1997. “Chromium as an Essential Nutrient for Humans.” Regulatory Toxicology and Pharmacology 26: S35–S41. - PubMed
-
- Beller, H. R. , Yang L., Varadharajan C., et al. 2014. “Divergent Aquifer Biogeochemical Systems Converge on Similar and Unexpected Cr(VI) Reduction Products.” Environmental Science & Technology 48: 10699–10706. - PubMed
-
- Blighe, K. , Rana S., and Lewis M.. 2024. “Enhancedvolcano: Publication‐Ready Volcano Plots With Enhanced Colouring and Labeling [Online].” Bioconductor—Open Source Software for Bioinformatics.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

