A set of plasmatic microRNA related to innate immune response highly predicts the onset of immune reconstitution inflammatory syndrome in tuberculosis co-infected HIV individuals (ANRS-12358 study)
- PMID: 40625756
- PMCID: PMC12231349
- DOI: 10.3389/fimmu.2025.1603338
A set of plasmatic microRNA related to innate immune response highly predicts the onset of immune reconstitution inflammatory syndrome in tuberculosis co-infected HIV individuals (ANRS-12358 study)
Abstract
Background: After initiation of combination antiretroviral treatment (cART), HIV-1/tuberculosis coinfected patients are at high risk of developing tuberculosis-associated immune reconstitution inflammatory syndrome (TB-IRIS). MicroRNAs, small molecules of approximately 22 nucleotides, which regulate post-transcriptional gene expression and their profile has been proposed as a biomarker for many diseases. We tested whether the microRNA profile could be a predictive biomarker for TB-IRIS.
Methods: Twenty-six selected microRNAs involved in the regulation of the innate immune response were investigated. Free plasmatic and microRNA-derived exosomes were measured by flow cytometry. The plasma from 74 HIV-1+TB+ individuals (35 IRIS and 39 non-IRIS) at the time of the diagnosis and before any treatment (baseline) of CAMELIA trial (ANRS1295-CIPRA KH001-DAIDS-ES ID10425); 15 HIV+TB- and 23 HIV-TB+, both naïve of any treatment; and 20 HIV-TB- individuals as controls were analysed.
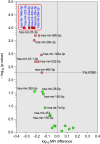
Results: At baseline, both IRIS and non-IRIS HIV+/TB+ individuals had similar demographic and clinical characteristics, including sex, age, body mass index, very low CD4+ cell counts (27 cells/mm3), and plasma HIV RNA load levels (5.76 log copies/ml). Twenty out of 26 plasmatic-microRNAs tested were no different between IRIS and controls. Twelve of the 26 tested microRNAs showed statistically significant differences between IRIS and non-IRIS patients (p-values ranging from p <0.05 to p <0.0001). Among these, five could discriminate between IRIS and non-IRIS individuals using ROC curve analysis (AUC scores ranging from 0.74 to 0.92). The combination of two (hsa-mir-29c-3p and hsa-mir-146a-5p) or three microRNAs (hsa-mir-29c-3p, hsa-mir-29a-3p, and hsa-mir-146a-5p) identified IRIS with 100% sensitivity and high specificity (95% and 97%, respectively).
Conclusion: The combination of at least two or three plasmatic microRNAs known to regulate inflammation and/or cytokine responses could be used as biomarkers to discriminate IRIS from non-IRIS in HIV-TB co-infected individuals at the time of diagnosis and prior to any treatment.
Keywords: HIV; IRIS; biomarkers; exosomes; microRNA; tuberculosis.
Copyright © 2025 Pean, Meng, Benichou, Srey, Dim, Borand, Marcy, Laureillard, Blanc, Cantaert, Madec, Weiss and Scott-Algara.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

