Developing a molecular diagnostic model for heatstroke-induced coagulopathy: a proteomics and metabolomics approach
- PMID: 40626023
- PMCID: PMC12231342
- DOI: 10.3389/fmolb.2025.1616073
Developing a molecular diagnostic model for heatstroke-induced coagulopathy: a proteomics and metabolomics approach
Abstract
Background: Heatstroke (HS) is becoming more concerning, with coagulopathy contributing to higher mortality. The aim of this study was to analyze the metabolomic and proteomic profiles associated with heatstroke-induced coagulopathy (HSIC) and to develop a molecular diagnostic model based on proteomic and metabolomic patterns.
Methods: This study included 41 HS patients from the Department of Critical Care Medicine at a comprehensive teaching hospital. Plasma proteins and metabolites from HSIC and non-heatstroke-induced coagulopathy (NHSIC) patients were compared using LC-MS/MS. Multivariate and univariate statistical analyses identified differentially expressed proteins (DEPs) and metabolites (DEMs). Functional annotation and pathway enrichment analyses were performed using the GO and KEGG databases, and machine learning models were developed using candidate proteins selected by LASSO and Boruta algorithms to diagnose HSIC. Finally, bioinformatic analysis was used to integrate the results of proteomics and metabolomics to find the potential mechanisms of HSIC.
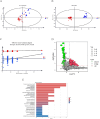
Results: A total of 41 patients participated in the study, with 11 cases in the HSIC group and 30 cases in the NHSIC group. Significant differences were observed between the groups in temperature, heart rate, white blood cell count, platelet count, liver function, coagulation markers, APACHE II score, and GCS score. Survival analysis revealed that the heatstroke group had a higher mortality risk. A total of 125 DEPs and 110 DEMs were identified, primarily enriched in energy regulation-related pathways and lipid and carbohydrate metabolism. Additionally, three optimal predictive models (AUC >0.9) were developed and validated for classifying HSIC from HS individuals based on proteomic patterns and machine learning, with the logistic regression model showing the best diagnostic performance (AUC = 0.979, sensitivity = 81.8%, specificity = 96.7%), highlighting lactate dehydrogenase A chain (LDHA), neutrophil gelatinase-associated lipocalin (NGAL), prothrombin and glucan-branching enzyme (GBE) as key predictors of HSIC.
Conclusion: The study uncovered critical metabolic and protein changes linked to heatstroke, highlighting the involvement of energy regulation, lipid metabolism, and carbohydrate metabolism. Building on these findings, an optimal machine learning diagnostic model was developed to boost the accuracy of HSIC diagnosis, integrating LDHA, NGAL, prothrombin, and GBE as key biomarkers.
Keywords: coagulopathy; heatstroke; machine learning; metabolomics; proteomics.
Copyright © 2025 Zeng, Lin, He, Zhong, Zhou, Deng, Zhang, Song and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Author Anonymous (2021). Update: Heat illness, active component, U.S. Armed Forces, 2020. MSMR 28 (4), 10–15. - PubMed
-
- Capozzi A., Riitano G., Recalchi S., Manganelli V., Costi R., Saccoliti F., et al. (2021). Effect of heparanase inhibitor on tissue factor overexpression in platelets and endothelial cells induced by anti-β2-GPI antibodies. J. Thromb. Haemost. 19 (9), 2302–2313. 10.1111/jth.15417 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

