Human pegivirus alters brain and blood immune and transcriptomic profiles of patients with Parkinson's disease
- PMID: 40626361
- PMCID: PMC12244338
- DOI: 10.1172/jci.insight.189988
Human pegivirus alters brain and blood immune and transcriptomic profiles of patients with Parkinson's disease
Abstract
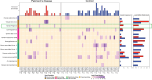
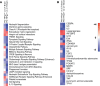
Parkinson's disease (PD) is a neurodegenerative disorder with both genetic and environmental factors contributing to pathogenesis. Viral infections are potential environmental triggers that influence PD pathology. Using ViroFind, an unbiased platform for whole virome sequencing, along with quantitative PCR (qPCR), we identified human pegivirus (HPgV) in 5 of 10 (50%) of PD brains, confirmed by IHC in 2 of 2 cases, suggesting an association with PD. All 14 age- and sex-matched controls were HPgV negative. HPgV-brain positive patients with PD showed increased neuropathology by Braak stage and Complexin-2 levels, while those positive in the blood had higher IGF-1 and lower pS65-ubiquitin, supporting disruption in metabolism or mitophagy in response to HPgV. RNA-Seq revealed altered immune signaling in HPgV-infected PD samples, including consistent suppression of IL-4 signaling in both the brain and blood. Longitudinal analysis of blood samples showed a genotype-dependent viral response, with HPgV titers correlating directly with IL-4 signaling in a LRRK2 genotype-dependent manner. YWHAB was a key hub gene in the LRRK2 genotypic response, which exhibited an altered relationship with immune-related factors, including NFKB1, ITPR2, and LRRK2 itself, in patients with PD who are positive for HPgV. These results suggest a role for HPgV in shaping PD pathology and highlight the complex interplay between viral infection, immunity, and neuropathogenesis.
Keywords: Genetic variation; Neurodegeneration; Neuroscience; Parkinson disease; Virology.
Conflict of interest statement
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

