A Method for Estimating Dynamic Functional Network Connectivity Gradients (dFNGs) From ICA Captures Smooth Inter-Network Modulation
- PMID: 40626410
- PMCID: PMC12235489
- DOI: 10.1002/hbm.70262
A Method for Estimating Dynamic Functional Network Connectivity Gradients (dFNGs) From ICA Captures Smooth Inter-Network Modulation
Abstract
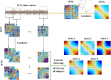
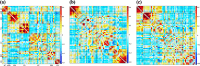
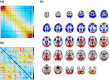
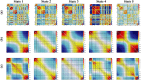
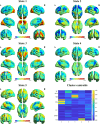
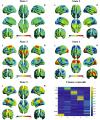
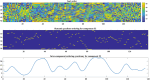
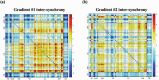
Dynamic functional network connectivity (dFNC) analysis is a widely used approach for studying brain function and offering insight into how brain networks evolve over time. Typically, dFNC studies utilize fixed spatial maps and evaluate transient changes in coupling among time courses estimated from independent component analysis (ICA). This manuscript presents a complementary approach that relaxes this assumption by spatially reordering the components dynamically at each time point to optimize for a smooth gradient in the FNC (i.e., a smooth gradient among ICA connectivity values). Several methods are presented to summarize dynamic FNC gradients (dFNGs) over time, starting with static FNC gradients (sFNGs), then exploring the reordering properties as well as the dynamics of the gradients themselves. We then apply this approach to a dataset of schizophrenia (SZ) patients and healthy controls (HCs). Functional dysconnectivity between different brain regions has been reported in SZ, yet the neural mechanisms behind it remain elusive. Using resting-state fMRI and ICA on a dataset consisting of 151 SZ patients and 160 age and gender-matched HCs, we extracted 53 intrinsic connectivity networks (ICNs) for each subject using a fully automated spatially constrained ICA approach. We develop several summaries of our functional network connectivity gradient analysis, both in a static sense, computed as the Pearson correlation coefficient between full time series, and a dynamic sense, computed using a sliding window approach followed by reordering based on the computed gradient, and evaluate group differences. Static connectivity analysis revealed significantly stronger connectivity between subcortical (SC), auditory (AUD), and visual (VIS) networks in patients, as well as hypoconnectivity in the sensorimotor (SM) network relative to controls. sFNG analysis highlighted distinctive clustering patterns in patients and HCs along cognitive control (CC)/default mode network (DMN), as well as SC/AUD/SM/cerebellar (CB) and VIS gradients. Furthermore, we observed significant differences in the sFNGs between groups in SC and CB domains. dFNG analysis suggested that SZ patients spend significantly more time in a SC/CB state based on the first gradient, while HCs favor the SM/DMN state. For the second gradient, however, patients exhibited significantly higher activity in CB domains, contrasting with HCs' DMN engagement. The gradient synchrony analysis conveyed more shifts between SM/SC networks and transmodal CC/DMN networks in patients. In addition, the dFNG coupling revealed distinct connectivity patterns between SC, SM, and CB domains in SZ patients compared to HCs. To recap, our results advance our understanding of brain network modulation by examining smooth connectivity trajectories. This provides a more complete spatiotemporal summary of the data, contributing to the growing body of current literature regarding the functional dysconnectivity in SZ patients. By employing dFNG, we highlight a new perspective to capture large-scale fluctuations across the brain while maintaining the convenience of brain networks and low-dimensional summary measures.
Keywords: dynamic functional network connectivity (dFNC); dynamic functional network connectivity gradient (dFNG); gradient; independent component analysis (ICA); schizophrenia.
© 2025 The Author(s). Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Update of
-
A method for estimating dynamic functional network connectivity gradients (dFNG) from ICA captures smooth inter-network modulation.bioRxiv [Preprint]. 2024 Jun 18:2024.03.06.583731. doi: 10.1101/2024.03.06.583731. bioRxiv. 2024. Update in: Hum Brain Mapp. 2025 Jul;46(10):e70262. doi: 10.1002/hbm.70262. PMID: 38559041 Free PMC article. Updated. Preprint.
Similar articles
-
Static and Dynamic Cross-Network Functional Connectivity Shows Elevated Entropy in Schizophrenia Patients.bioRxiv [Preprint]. 2024 Jun 17:2024.06.15.599084. doi: 10.1101/2024.06.15.599084. bioRxiv. 2024. Update in: Hum Brain Mapp. 2025 Feb 01;46(2):e70134. doi: 10.1002/hbm.70134. PMID: 38948857 Free PMC article. Updated. Preprint.
-
A method for estimating dynamic functional network connectivity gradients (dFNG) from ICA captures smooth inter-network modulation.bioRxiv [Preprint]. 2024 Jun 18:2024.03.06.583731. doi: 10.1101/2024.03.06.583731. bioRxiv. 2024. Update in: Hum Brain Mapp. 2025 Jul;46(10):e70262. doi: 10.1002/hbm.70262. PMID: 38559041 Free PMC article. Updated. Preprint.
-
Alterations in Gray Matter Structure Linked to Frequency-Specific Cortico-Subcortical Connectivity in Schizophrenia via Multimodal Data Fusion.Neuroinformatics. 2025 Apr 26;23(2):31. doi: 10.1007/s12021-025-09728-3. Neuroinformatics. 2025. PMID: 40285903
-
Interventions for central serous chorioretinopathy: a network meta-analysis.Cochrane Database Syst Rev. 2025 Jun 16;6(6):CD011841. doi: 10.1002/14651858.CD011841.pub3. Cochrane Database Syst Rev. 2025. PMID: 40522203
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
Unraveling the Neural Landscape of Mental Disorders using Double Functional Independent Primitives (dFIPs).bioRxiv [Preprint]. 2024 Aug 2:2024.08.01.606076. doi: 10.1101/2024.08.01.606076. bioRxiv. 2024. Update in: Biol Psychiatry Cogn Neurosci Neuroimaging. 2025 Apr 11:S2451-9022(25)00129-6. doi: 10.1016/j.bpsc.2025.03.015. PMID: 39131299 Free PMC article. Updated. Preprint.
-
Static and Dynamic Cross-Network Functional Connectivity Shows Elevated Entropy in Schizophrenia Patients.bioRxiv [Preprint]. 2024 Jun 17:2024.06.15.599084. doi: 10.1101/2024.06.15.599084. bioRxiv. 2024. Update in: Hum Brain Mapp. 2025 Feb 01;46(2):e70134. doi: 10.1002/hbm.70134. PMID: 38948857 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

