The ribosomal RNA transcription landscapes of Plasmodium falciparum and related apicomplexan parasites
- PMID: 40626563
- PMCID: PMC12235521
- DOI: 10.1093/nar/gkaf641
The ribosomal RNA transcription landscapes of Plasmodium falciparum and related apicomplexan parasites
Abstract
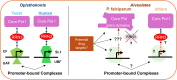
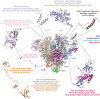
Ribosome biogenesis is essential for the rapid proliferation and life cycle transitions of Plasmodium falciparum, the causative agent of malaria. In eukaryotes, ribosomal RNA synthesis is carried out by RNA polymerase I (Pol I), highly specialized transcriptional machinery. This review provides a comparative analysis of Pol I transcription apparatus in yeast and humans, serving as a reference framework to examine its evolutionary divergence in P. falciparum and related apicomplexans and alveolates. Bioinformatic analyses revealed that some of these organisms lack any identifiable homologues or orthologs of several canonical eukaryotic transcription factors essential for Pol I-mediated transcription, including initiation factor RRN3, activator UBF, and all specific subunits of the promoter recognition complexes. Interestingly, the parasite retains core Pol I subunits, incorporating unique parasite-specific structural domains characterized through AI-based protein complex modeling of P. falciparum Pol I. These adaptations may compensate for the absence of traditional regulatory factors, enabling the parasite to employ distinct mechanisms for promoter recognition and transcription initiation. The substantial differences between parasite and host Pol I transcription machinery create potential targets for therapeutic intervention with parasite-specific elements representing potential drug targets. By integrating evolutionary, structural, and functional perspectives, this review advances our understanding of Pol I transcription in alveolates and its implications for the development of novel antimalarial strategies.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

