The miR-302 cluster-IRFs-IRF1AS axis regulates influenza A virus replication in a species-specific manner
- PMID: 40626712
- PMCID: PMC12345190
- DOI: 10.1128/mbio.01375-25
The miR-302 cluster-IRFs-IRF1AS axis regulates influenza A virus replication in a species-specific manner
Abstract
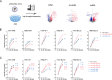
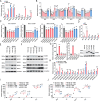
Non-coding RNAs are crucial orchestrators in the intricate dance between viruses and host cells, among which the expression and function of enhancer RNAs (eRNAs) during influenza virus infection remain largely unexplored. This study utilized whole transcriptome high-throughput sequencing to investigate the molecular mechanisms underpinning the species-specific regulation of influenza virus replication by the miR-302 cluster-IRFs-IRF1AS axis both in vivo and in vitro. Mechanistically, the CTNNB1-induced miR-302 cluster targeted various interferon regulatory factors (mainly IRF1 and IRF2) with varying affinities and silencing efficiencies, except for miR-302e and miR-302f. Furthermore, miR-302 cluster-IRFs triggered the induction of interferon-induced hub genes and hub lncRNAs defined through weighted gene co-expression network analysis. Importantly, the intricate interplay between IRFs, direct targets of the miR-302 cluster, and IRF1AS, an indirect target, in terms of gene loci and transcriptional regulation reveals a crosstalk in the miR-302 cluster-IRFs-IRF1AS axis. That is, on the one hand, IRF1 and IRF7 bind to the promoter of IRF1AS to promote the transcription of eRNAs. On the other hand, IRF1AS functions as an enhancer cluster that orchestrates and cis-regulates the transcription of IRF1, thereby rapidly amplifying the antiviral immune response initiated by miR-302 cluster-IRFs. In conclusion, we have unveiled a novel regulatory network governed by the miR-302 cluster-IRFs-IRF1AS, offering fresh perspectives on immune regulatory mechanisms.
Importance: Non-coding RNAs play a crucial role in regulating the three-dimensional structure of chromatin. They influence gene expression through various mechanisms and thereby contribute to the onset and progression of influenza A virus pathogenicity. Our comprehensive whole transcriptome sequencing analysis reveals a novel finding: the species-specific regulation of influenza virus replication by the miR-302 cluster-IRFs-IRF1AS axis. Our findings indicate that the miR-302 cluster-IRFs axis facilitates the transcription of key hub genes and hub lncRNAs, most of which significantly inhibit influenza virus replication. Notably, the downstream IRF1AS assembles into an enhancer cluster, orchestrating and cis-regulating the transcription of IRF1 to activate the interferon system. This investigation enhances our understanding of the regulatory network underlying viral infections and offers novel insights into immune regulatory mechanisms.
Keywords: IRFs-IRF1AS; enhancer RNAs; enhancer cluster; influenza A virus; miR-302 cluster-IRFs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection.J Virol. 2017 Mar 29;91(8):e02388-16. doi: 10.1128/JVI.02388-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148804 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Functional comparison of tilapia interferon regulatory factor (IRF) 1 and IRF11 in type I interferon-mediated antiviral responses.Int J Biol Macromol. 2025 Jul;318(Pt 2):145077. doi: 10.1016/j.ijbiomac.2025.145077. Epub 2025 Jun 8. Int J Biol Macromol. 2025. PMID: 40494468
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
References
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
-
- Eisfeld AJ, Biswas A, Guan L, Gu C, Maemura T, Trifkovic S, Wang T, Babujee L, Dahn R, Halfmann PJ, Barnhardt T, Neumann G, Suzuki Y, Thompson A, Swinford AK, Dimitrov KM, Poulsen K, Kawaoka Y. 2024. Pathogenicity and transmissibility of bovine H5N1 influenza virus. Nature 633:426–432. doi: 10.1038/s41586-024-07766-6 - DOI - PMC - PubMed
-
- Guan L, Eisfeld AJ, Pattinson D, Gu C, Biswas A, Maemura T, Trifkovic S, Babujee L, Presler R Jr, Dahn R, Halfmann PJ, Barnhardt T, Neumann G, Thompson A, Swinford AK, Dimitrov KM, Poulsen K, Kawaoka Y. 2024. Cow's milk containing avian influenza A(H5N1) virus - heat inactivation and infectivity in mice. N Engl J Med 391:87–90. doi: 10.1056/NEJMc2405495 - DOI - PubMed
-
- Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus A, Fouchier RAM. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
