LINC2781 enhances antiviral immunity against coxsackievirus B5 infection by activating the JAK-STAT pathway and blocking G3BP2-mediated STAT1 degradation
- PMID: 40626722
- PMCID: PMC12306156
- DOI: 10.1128/msphere.00062-25
LINC2781 enhances antiviral immunity against coxsackievirus B5 infection by activating the JAK-STAT pathway and blocking G3BP2-mediated STAT1 degradation
Abstract
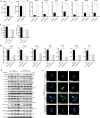
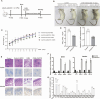
Coxsackievirus B5 (CVB5) is a primary causative agent of hand, foot, and mouth disease (HFMD), and some cases are also associated with severe complications through invasion of the central nervous system, resulting in death. Currently, there are no specific antiviral drugs or effective vaccines available for CVB5. Long non-coding RNAs (lncRNAs) have been shown to play significant roles in various diseases. In our research, we identified a novel lncRNA, LINC2781, which is significantly upregulated during CVB5 infection of SH-SY5Y cells. Characteristic analysis showed that LINC2781 is mainly located in the cytoplasm of infected cells, with significantly higher expression in the intestines and the spleen of CVB5-infected mice. Functional studies revealed that LINC2781 activates the JAK-STAT pathway via STAT1, promoting the expression of interferon-stimulated genes (ISGs) and inhibiting CVB5 replication. Mechanistically, LINC2781 directly binds to GTPase-activating protein SH3 domain-binding protein 2 (G3BP2), preventing G3BP2-mediated degradation of STAT1 through ubiquitination. In vivo, LINC2781 was shown to reduce the susceptibility of BALB/c mice to viral infection and alleviate viral-induced damage in both the intestines and the spleen. Clinical samples further confirmed a strong correlation between the expression of LINC2781 and CVB5 infection. Our findings demonstrate that CVB5-induced LINC2781 enhances STAT1 activation by blocking the suppressive effects of G3BP2 on immune responses, providing a potential foundation for developing antiviral therapies targeting the lncRNA.
Importance: We investigate the role of lncRNA in virus-host interactions and identify a novel cytoplasmic lncRNA, LINC2781, whose expression is upregulated following CVB5 infection. LINC2781 specifically binds to G3BP2, preventing G3BP2 from degrading STAT1, thereby activating the JAK-STAT pathway, promoting the expression of ISGs, and ultimately inhibiting viral replication. Meanwhile, a strong correlation exists between the expression of LINC2781 and CVB5 infection in cells and clinical samples.
Keywords: LINC2781; GTPase-activating protein SH3 domain-binding protein 2 (G3BP2); STAT1; coxsackievirus B5 (CVB5); long non-coding RNAs (lncRNAs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
miR-216a-3p alleviates primary Sjögren's syndrome by regulating the STAT1/JAK signaling pathway.Biochem Biophys Res Commun. 2025 Apr 12;758:151647. doi: 10.1016/j.bbrc.2025.151647. Epub 2025 Mar 17. Biochem Biophys Res Commun. 2025. PMID: 40121969
-
The AhR regulates IFN-induced immune checkpoints in lung cancer cells through HNRNPH1, an RNA-binding protein, and INCR1, a novel long non-coding RNA.J Biol Chem. 2025 Jul;301(7):110316. doi: 10.1016/j.jbc.2025.110316. Epub 2025 May 29. J Biol Chem. 2025. PMID: 40449595 Free PMC article.
-
Sex differences in mitochondrial gene expression during viral myocarditis.Biol Sex Differ. 2024 Dec 18;15(1):104. doi: 10.1186/s13293-024-00678-0. Biol Sex Differ. 2024. PMID: 39696682 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
