NOTCH1 reverses immune suppression in small cell lung cancer through reactivation of STING
- PMID: 40627448
- PMCID: PMC12435836
- DOI: 10.1172/JCI185423
NOTCH1 reverses immune suppression in small cell lung cancer through reactivation of STING
Abstract
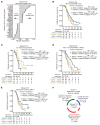
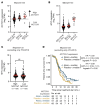
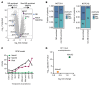
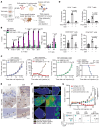
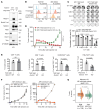
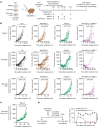
Downregulation of antigen presentation and lack of immune infiltration are defining features of small cell lung cancer (SCLC), limiting response to immune checkpoint blockade (ICB). While a high-MHC class I, immune-inflamed subset benefits from ICB, underlying mechanisms of immune response in SCLC have yet to be elucidated. Here we show that in the IMpower133 clinical trial, high, but not low, NOTCH1 expression was significantly associated with longer survival with the addition of ICB to chemotherapy among approximately 80% of SCLC patients with NE-enriched tumors (ASCL1-enriched, HR 0.39, P = 0.0012; NEUROD1-enriched, HR 0.44, P = 0.024). Overexpression or pharmacologic activation of NOTCH1 in ASCL1 and NEUROD1 SCLC cell lines dramatically upregulated MHC class I through epigenetic reactivation of STING. In syngeneic mouse models, Notch1 activation reprogrammed SCLC tumors from immune-excluded to immune-inflamed, facilitating durable, complete responses with ICB combined with a STING agonist. STING1 expression was significantly enriched in high- compared with low-NOTCH1-expressing tumors in IMpower133, validating our proposed mechanism. Our data reveal a previously undiscovered role for NOTCH1 as a critical driver of SCLC immunogenicity and a potential predictive biomarker for ICB in SCLC. NOTCH1 activation may be a therapeutic strategy to unleash antitumor immune responses in SCLC and other neuroendocrine cancers in which NOTCH1 is typically suppressed.
Keywords: Antigen; Biomarkers; Cancer immunotherapy; Cell biology; Immunology; Oncology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

