Activity-Based Tracking of Glycan Turnover in Microbiomes
- PMID: 40628650
- PMCID: PMC12291463
- DOI: 10.1021/jacs.5c07546
Activity-Based Tracking of Glycan Turnover in Microbiomes
Abstract
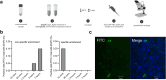
Glycans shape microbiomes in the ocean and the gut, driving key steps in the global carbon cycle and human health. Yet, our ability to track microbial glycan turnover across microbiomes is limited, as identifying active degraders without prior genomic knowledge remains a key challenge. Here, we introduce an activity-based fluorescence resonance energy transfer (FRET) probe that enables direct visualization and quantification of glycan metabolism in complex microbial communities. As a proof of concept, we investigated α-mannan degradation, a prominent polysaccharide in algal blooms. Using automated glycan assembly, we synthesized a mannan hexasaccharide bearing a fluorescein-rhodamine FRET pair. The probe was validated using a recombinantly expressed endo-α-mannanase (GH76) from Salegentibacter sp. Hel_I_6. It was shown to function in cell lysates, pure cultures, and complex microbiomes (via plate assays and microscopy). This probe enabled spatiotemporal visualization of in situ α-mannan turnover in a marine microbiome. Glycan FRET probes are versatile tools for tracking glycan degradation across biological scales from single enzymes to microbiomes.
Figures




Similar articles
-
Stronger together: harnessing natural algal communities as potential probiotics for inhibition of aquaculture pathogens.Microbiol Spectr. 2025 Jul;13(7):e0042125. doi: 10.1128/spectrum.00421-25. Epub 2025 May 21. Microbiol Spectr. 2025. PMID: 40396728 Free PMC article.
-
A bloom of a single bacterium shapes the microbiome during outdoor diatom cultivation collapse.mSystems. 2025 Jun 17;10(6):e0037525. doi: 10.1128/msystems.00375-25. Epub 2025 May 14. mSystems. 2025. PMID: 40366134 Free PMC article.
-
A tale of two vineyards: parsing site-specific differences in bacterial and fungal communities of wine grapes from proximal vineyards and their changes during processing in a single winery.Appl Environ Microbiol. 2025 Jun 18;91(6):e0052625. doi: 10.1128/aem.00526-25. Epub 2025 May 5. Appl Environ Microbiol. 2025. PMID: 40323100 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

