Pex3 promotes formation of peroxisome-peroxisome and peroxisome-lipid droplet contact sites
- PMID: 40628847
- PMCID: PMC12238565
- DOI: 10.1038/s41598-025-07934-2
Pex3 promotes formation of peroxisome-peroxisome and peroxisome-lipid droplet contact sites
Abstract
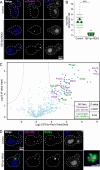
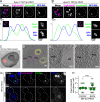
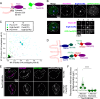
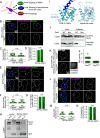
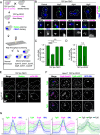
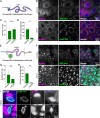
Peroxisomes are ubiquitous organelles that mediate central metabolic functions, such as fatty acid β-oxidation, as well as diverse tissue- and organism-specific processes. Membrane contact sites, regions of close apposition with other organelles for direct communication, are central to several aspects of their life cycle. Pex3 is a conserved multifunctional peroxisomal transmembrane protein that is involved in the insertion of peroxisomal membrane proteins, in pexophagy, and in the formation of membrane contact sites. Here, we show that high Pex3 levels in Saccharomyces cerevisiae induce the formation of peroxisome clusters surrounded by lipid droplets, mediated by peroxisome-peroxisome and peroxisome-lipid droplet contact sites. This clustering occurs independently of Pex3 partners in other processes Pex19, Inp1, and Atg36. The cytosolic domain of Pex3 binds peroxisomes, suggesting a direct role in homotypic contact site formation. Lipid droplet-peroxisome contact sites require the lipid droplet-localized triacylglycerol lipase Tgl4, which is enriched at this interface along with other lipases. Pex3 overexpression in Drosophila melanogaster similarly alters peroxisome and lipid droplet morphology and promotes contact site formation. Together, our results offer novel molecular insights into homotypic peroxisome contact sites and peroxisome-lipid droplet contact sites across species.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- Wanders, R. J. A., Baes, M., Ribeiro, D., Ferdinandusse, S. & Waterham, H. R. The physiological functions of human peroxisomes. Physiol. Rev.103, 957–1024 (2023). - PubMed
-
- Eisenberg-Bord, M., Shai, N., Schuldiner, M. & Bohnert, M. A tether is a tether is a tether: Tethering at membrane contact sites. Dev. Cell39, 395–409 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

