Integrated multi-omics of feces, plasma and urine can describe and differentiate pediatric active Crohn's Disease from remission
- PMID: 40629054
- PMCID: PMC12238252
- DOI: 10.1038/s43856-025-00984-7
Integrated multi-omics of feces, plasma and urine can describe and differentiate pediatric active Crohn's Disease from remission
Abstract
Background: This study aimed to obtain a holistic view of remission in pediatric Crohn's Disease (CD) by integrating six omics datasets from three anatomical compartments.
Methods: Patients with fecal calprotectin below 250 mg/kg were considered in remission (n = 27), above 250 mg/kg as having active disease (n = 31). Proteome and microbiomes (fungi and bacteria) were analyzed in feces. Metabolomes in feces, urine, and plasma. Datasets were integrated into a multi-omics model.
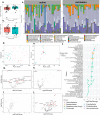
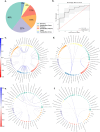
Results: The use of individual datasets shows multiple differences between remission and active disease. Integration yielded a good model (AUC of 0.8) predicting remission. The most important features in this model are fecal bacteria (40%), fecal metabolites (22%), fecal proteins (16%), plasma metabolites (12%), fecal fungi (6%), and urine metabolites (4%). The interactome reveals Ruminococcaceae and Faecalibacterium as key players, with a correlation between antifungal urine hydroxyphenyllactic acid and fecal fungi. Pathway analysis shows an association of purine metabolism with remission, independent of thiopurine use. Changes in purine metabolism are confirmed in a pediatric CD public dataset.
Conclusion: The pathways and correlations identified as playing a role in remission may remain undetectable if individual omics datasets or single anatomical compartments are used, highlighting the need for a holistic approach that integrates multiple datasets from multiple anatomical compartments.
Plain language summary
Crohn’s disease is a severe inflammation of the intestine, characterized by periods of remission and active disease. Intestinal bacteria and fungi, and compounds produced by these organisms and the patient, all play a role in the development of Crohn’s disease. Because patients were able to submit feces, blood, and urine, we could determine how these factors interact and are different between patients with active disease and in remission. This was the first time all these different factors could be measured in the same patient, allowing a holistic and molecular look at patients with Crohn’s disease. We identified key bacteria and pathways that are important in the remission of Crohn’s disease. Our research may point at new directions for the treatment of this severe disease.
© 2025. The Author(s).
Figures



References
LinkOut - more resources
Full Text Sources

