hafoe: an interactive tool for the analysis of chimeric AAV libraries after random mutagenesis
- PMID: 40629114
- PMCID: PMC12518119
- DOI: 10.1038/s41434-025-00548-3
hafoe: an interactive tool for the analysis of chimeric AAV libraries after random mutagenesis
Abstract
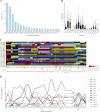
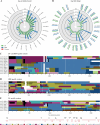
Naturally occurring adeno-associated viruses (AAVs) are an integral part of gene therapy, yet engineering novel AAV variants is necessary to expand targetable tissues and treatable diseases. Directed evolution, particularly through DNA shuffling of the capsid genes of wild-type AAV serotypes, is a widely employed strategy to generate novel chimeric variants with desired properties. Yet, the computational analysis of such chimeric sequences presents challenges. We introduce hafoe, a novel computational tool designed for the exploratory analysis of chimeric AAV libraries, which does not require extensive bioinformatics expertise. hafoe accurately deciphers the serotype composition and enrichment patterns of chimeric AAV variants across different tissues. Validation against synthetic datasets demonstrates that hafoe identifies parental serotype compositions with an accuracy of 96.3% to 97.5%. Additionally, we engineered chimeric AAV capsid libraries and screened novel AAV variants for tropism to human dermal fibroblasts and dendritic cells, as well as canine muscle, and liver tissues. Using hafoe we identified and characterized enriched AAV variants in these tissues for potential use in gene therapy and vaccine development. Overall, hafoe can provide valuable insights that may further support the rational design of AAV vectors based on parental serotype and sequence preferences of the capsid genes in target tissues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Harvard University has filed for patent protection on DC and HDF-enriched AAV variants described herein, and EA, DM, and GMC are named as co-inventors on the patent. At the time of study, EA performed consulting services, and RH and ND were employees and held equity in Rejuvenate Bio. DM and DBT are full-time employees and hold equity in Medici Therapeutics, Inc. For a complete list of GMC's financial interests, see http://arep.med.harvard.edu/gmc/tech.html . The remaining authors declare no competing interests. Ethical approval: All procedures involving Beagle dogs in this study were conducted in accordance with the USDA Animal Welfare Act (9 CFR, Parts 1, 2, and 3) and the Guide for Care and Use of Laboratory Animals (ILAR publication, 2011). The protocols were approved by the institutional ethical review board at Absorption Systems California, LLC (ASC), and conducted under the authority of the Project License issued by ASC’s compliance office. The animals were housed in compliance with ethical standards, provided with a certified laboratory diet (5007 laboratory canine diet from LabDiet), and had access to water ad libitum. The study followed the Standard Operating Procedures established at ASC, ensuring a high standard of care and scientific integrity throughout the experiment.
Figures




References
-
- Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–72. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

