Modulating ovarian cancer progression through FDX1-driven autophagy
- PMID: 40629130
- PMCID: PMC12238385
- DOI: 10.1038/s41698-025-00994-7
Modulating ovarian cancer progression through FDX1-driven autophagy
Abstract
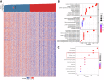
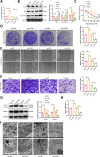
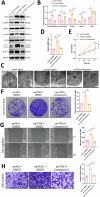
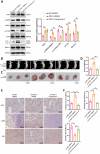
Ferredoxin 1 (FDX1) emerges as a crucial regulator of autophagy and copper metabolism in ovarian cancer (OC), as revealed by this investigation. Predominantly localized to the cytoplasm and mitochondria, FDX1 coordinates autophagic activity by modulating the AMPK and mTOR signaling pathways. Its role extends to preserving mitochondrial integrity and facilitating sulfation of DLAT/DLST, ensuring effective autophagic flux. Knockdown of FDX1 disrupts these processes, exacerbating mitochondrial dysfunction. In vivo studies further demonstrate that overexpressing FDX1, combined with Compound C treatment, markedly inhibits tumor growth and Ki67 expression. These results position FDX1 as a promising target for therapeutic strategies aimed at exploiting autophagy to hinder OC progression.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
p53 enhances elesclomol-Cu-induced cuproptosis in hepatocellular carcinoma via FDXR-mediated FDX1 upregulation.Front Oncol. 2025 Jun 24;15:1584811. doi: 10.3389/fonc.2025.1584811. eCollection 2025. Front Oncol. 2025. PMID: 40630211 Free PMC article.
-
Cuproptosis: a novel therapeutic mechanism in lung cancer.Cancer Cell Int. 2025 Jun 24;25(1):231. doi: 10.1186/s12935-025-03864-1. Cancer Cell Int. 2025. PMID: 40555995 Free PMC article. Review.
-
Modulation of Cuproptosis Pathway Genes (DLAT, FDX1) and Antioxidant Enzyme Activities in Obese Mice in Response to Quercetin and Calorie Restriction.DNA Cell Biol. 2025 Jul;44(7):370-379. doi: 10.1089/dna.2025.0005. Epub 2025 May 12. DNA Cell Biol. 2025. PMID: 40354319
-
miR-210 Regulates Autophagy Through the AMPK/mTOR Signaling Pathway, Reduces Neuronal Cell Death and Inflammatory Responses, and Enhances Functional Recovery Following Cerebral Hemorrhage in Mice.Neurochem Res. 2025 Jun 5;50(3):180. doi: 10.1007/s11064-025-04434-7. Neurochem Res. 2025. PMID: 40471451 Free PMC article.
-
Topotecan, pegylated liposomal doxorubicin hydrochloride and paclitaxel for second-line or subsequent treatment of advanced ovarian cancer: a systematic review and economic evaluation.Health Technol Assess. 2006 Mar;10(9):1-132. iii-iv. doi: 10.3310/hta10090. Health Technol Assess. 2006. PMID: 16545208
References
LinkOut - more resources
Full Text Sources
Miscellaneous

