FPGA acceleration of GWAS permutation testing
- PMID: 40630501
- PMCID: PMC12237511
- DOI: 10.1093/bioadv/vbaf145
FPGA acceleration of GWAS permutation testing
Abstract
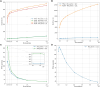
Genome-wide association studies (GWASs) analyse genetic variation across many individuals to identify single-nucleotide polymorphisms (SNPs) associated with complex traits. They typically include millions of SNPs from thousands of individuals, creating a multiple testing problem where the probability of false associations increases with the number of SNPs tested. While permutation testing provides accurate control of false positive rates, it is computationally expensive and slow for large datasets. This research presents an FPGA-based tool designed for cloud deployment on AWS EC2 instances that significantly accelerates GWAS permutation testing for continuous phenotypes. The tool implements two algorithms: maxT and adaptive permutation testing. Performance comparisons using a breast cancer dataset (13.7 million SNPs from 3652 individuals) showed large speedups over PLINK running on 40 CPU cores. For 1000 maxT permutations, the FPGA tool completed analysis in 22 min versus PLINK's 7 days. For 100 million adaptive permutations, FPGA required 325 min compared to PLINK's 8.5 days. The tool handled 700 million adaptive permutations in 33 h-a workload which would require over a month for CPU-based analysis. FPGA solution provides accessible, order-of-magnitude performance improvements without requiring FPGA expertise or dedicated cluster access.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare that there are no conflicts of interest related to this work. No competing financial, professional, or personal relationships influenced the design, execution, analysis, or interpretation of the study.
Figures


References
-
- Dudoit S, van der Laan MJ. Multiple Testing Procedures with Applications to Genomics. Springer Series in Statistics. New York: Springer, 2008.
-
- Goeman JJ, Solari A. Multiple hypothesis testing in genomics. Statistics in Medicine 2014;33:1946–78. - PubMed
-
- Gundlach S, Kässens JC, Wienbrandt L. Genome-wide association interaction studies with MB-MDR AND maxt multiple testing correction on FPGAs. Procedia Comput Sci 2016;80:639–49. 10.1016/j.procs.2016.05.354 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

