Differential extracellular matrix proteomic signatures in colorectal tumors from Appalachian and non-Appalachian patients
- PMID: 40630709
- PMCID: PMC12235701
- DOI: 10.3892/ol.2025.15159
Differential extracellular matrix proteomic signatures in colorectal tumors from Appalachian and non-Appalachian patients
Abstract
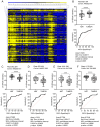
Emerging evidence reports that regulation of the extracellular matrix influences the progression of colorectal cancer (CRC). The present study investigated regulation of the extracellular matrix proteome in colorectal malignancy within a high-risk Appalachian population compared with non-Appalachian populations. A targeted mass spectrometry imaging proteomic method directed at collagen regulation was used. Tissue microarrays (TMAs) comprising of matched CRC with adjacent normal to tumor (NAT) from 45 patients were constructed into 86 samples to evaluate the extracellular matrix proteome (ECM). A total of five specific peaks were discovered to differ between NAT and tumor with high sensitivity and specificity by receiver operating characteristic (AUROC) ≥0.7, Wilson/Brown P<0.0002. Evaluation of patient TMA cores showed increased levels of combined ECM peptides in advanced stage Appalachian CRC (III + IV) compared with early staged CRC (I + II) (AUROC 0.8595; 95% confidence interval, 0.8190-0.8999; Wilson/Brown P<1.0×10-15), contrasting with the non-Appalachian tumors, which showed a decreased ability to discriminate between early and late stage (AUROC 0.6618; 95% confidence interval, 0.6126-0.7110; Wilson/Brown P<1.0×10-9). Comparison of advanced stage CRCs between Appalachian and non-Appalachian populations showed high sensitivity and specificity in distinguishing the populations (AUROC 0.7612; 95% confidence interval, 0.7109-0.8114; Wilson/Brown P<3.0×10-15). History of smoking, sex and tumor origin location did not show significant ability to distinguish by AUROC. A combination of high mass resolution, high mass accuracy spatial proteomics and sequencing proteomics by liquid chromatography coupled to tandem mass spectrometry revealed that fibrillar collagens were spatially regulated within the CRC tumor microenvironment. Fibrillar collagen post-translational modifications of hydroxylated proline revealed distinct spatial separation based on the presence of a number of hydroxylated proline sites. The present study highlighted that the targeted mass spectrometry imaging of the ECM proteome may provide new insight and novel predictive tools for understanding CRC, particularly among Appalachian patients.
Keywords: cancer; collagen; colorectal cancer; extracellular matrix; imaging; mass spectrometry imaging; spatial proteomics.
Copyright: © 2025 Sougiannis et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Nissen NI, Kehlet S, Boisen MK, Liljefors M, Jensen C, Johansen AZ, Johansen JS, Erler JT, Karsdal M, Mortensen JH, et al. Prognostic value of blood-based fibrosis biomarkers in patients with metastatic colorectal cancer receiving chemotherapy and bevacizumab. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-020-79608-0. - DOI - PMC - PubMed
-
- Iacovelli R, Pietrantonio F, Maggi C, de Braud F, Di Bartolomeo M. Combination or single-agent chemotherapy as adjuvant treatment of gastric cancer: A systematic review and meta-analysis of published trials. Crit Rev Oncol Hematol. 2016;98:24–28. doi: 10.1016/j.critrevonc.2015.09.002. - DOI - PubMed
-
- Iacovelli R, Pietrantonio F, Palazzo A, Maggi C, Ricchini F, de Braud F, Di Bartolomeo M. Incidence and relative risk of grade 3 and 4 diarrhoea in patients treated with capecitabine or 5-fluorouracil: A meta-analysis of published trials. Br J Clin Pharmacol. 2014;78:1228–1237. doi: 10.1111/bcp.12449. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
